3A8M
 
 | | Crystal structure of Nitrile Hydratase mutant Y72F complexed with Trimethylacetonitrile | | Descriptor: | 2,2-dimethylpropanenitrile, FE (III) ION, MAGNESIUM ION, ... | | Authors: | Yamanaka, Y, Hashimoto, K, Ohtaki, A, Noguchi, K, Yohda, M, Odaka, M. | | Deposit date: | 2009-10-06 | | Release date: | 2010-04-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Kinetic and structural studies on roles of the serine ligand and a strictly conserved tyrosine residue in nitrile hydratase
J.Biol.Inorg.Chem., 15, 2010
|
|
4QHP
 
 | | Crystal structure of Aminopeptidase N in complex with the phosphinic dipeptide analogue LL-(R,S)-hPheP[CH2]Phe(4-CH2NH2) | | Descriptor: | (2R)-2-[4-(aminomethyl)benzyl]-3-[(R)-[(1R)-1-amino-3-phenylpropyl](hydroxy)phosphoryl]propanoic acid, (2S)-2-[4-(aminomethyl)benzyl]-3-[(R)-[(1R)-1-amino-3-phenylpropyl](hydroxy)phosphoryl]propanoic acid, Aminopeptidase N, ... | | Authors: | Nocek, B, Joachimiak, A. | | Deposit date: | 2014-05-28 | | Release date: | 2014-09-24 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-guided, single-point modifications in the phosphinic dipeptide structure yield highly potent and selective inhibitors of neutral aminopeptidases.
J.Med.Chem., 57, 2014
|
|
8PTE
 
 | | Polyoxidovanadate interaction with proteins: crystal structure of lysozyme bound to octadecavanadate ion (structure B) | | Descriptor: | Lysozyme C, NITRATE ION, Polyoxidovanadate complex, ... | | Authors: | Tito, G, Ferraro, G, Merlino, A. | | Deposit date: | 2023-07-14 | | Release date: | 2024-01-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | Stabilization and Binding of [V 4 O 12 ] 4- and Unprecedented [V 20 O 54 (NO 3 )] n- to Lysozyme upon Loss of Ligands and Oxidation of the Potential Drug V IV O(acetylacetonato) 2.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
3A8L
 
 | | Crystal structure of photo-activation state of Nitrile Hydratase mutant S113A | | Descriptor: | FE (III) ION, Nitrile hydratase subunit alpha, Nitrile hydratase subunit beta | | Authors: | Yamanaka, Y, Hashimoto, K, Ohtaki, A, Noguchi, K, Yohda, M, Odaka, M. | | Deposit date: | 2009-10-06 | | Release date: | 2010-04-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Kinetic and structural studies on roles of the serine ligand and a strictly conserved tyrosine residue in nitrile hydratase
J.Biol.Inorg.Chem., 15, 2010
|
|
3QTT
 
 | | Crystal Structure of Pantoate-beta-alanine Ligase from Francisella tularensis Complexed with Beta-gamma ATP and Beta-alanine | | Descriptor: | BETA-ALANINE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Maltseva, N, Kim, Y, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-02-23 | | Release date: | 2011-03-23 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.599 Å) | | Cite: | Crystal Structure of Pantoate-beta-alanine Ligase from Francisella tularensis Complexed with Beta-gamma ATP and Beta-alanine.
To be Published
|
|
1J2E
 
 | | Crystal structure of Human Dipeptidyl peptidase IV | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase IV | | Authors: | Hiramatsu, H, Kyono, K, Higashiyama, Y, Fukushima, C, Shima, H, Sugiyama, S, Inaka, K, Yamamoto, A, Shimizu, R. | | Deposit date: | 2002-12-30 | | Release date: | 2003-12-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure and function of human dipeptidyl peptidase IV, possessing a unique eight-bladed beta-propeller fold.
Biochem.Biophys.Res.Commun., 302, 2003
|
|
3A8O
 
 | | Crystal structure of Nitrile Hydratase complexed with Trimethylacetamide | | Descriptor: | 2,2-dimethylpropanamide, FE (III) ION, Nitrile hydratase subunit alpha, ... | | Authors: | Yamanaka, Y, Hashimoto, K, Ohtaki, A, Noguchi, K, Yohda, M, Odaka, M. | | Deposit date: | 2009-10-07 | | Release date: | 2010-04-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Kinetic and structural studies on roles of the serine ligand and a strictly conserved tyrosine residue in nitrile hydratase
J.Biol.Inorg.Chem., 15, 2010
|
|
6YB7
 
 | | SARS-CoV-2 main protease with unliganded active site (2019-nCoV, coronavirus disease 2019, COVID-19). | | Descriptor: | 3C-like proteinase, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE | | Authors: | Owen, C.D, Lukacik, P, Strain-Damerell, C.M, Douangamath, A, Powell, A.J, Fearon, D, Brandao-Neto, J, Crawshaw, A.D, Aragao, D, Williams, M, Flaig, R, Hall, D.R, McAuley, K.E, Mazzorana, M, Stuart, D.I, von Delft, F, Walsh, M.A. | | Deposit date: | 2020-03-16 | | Release date: | 2020-03-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | COVID-19 main protease with unliganded active site
To Be Published
|
|
7OML
 
 | |
7OLH
 
 | |
6XPC
 
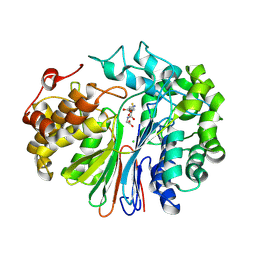 | | Structure of human GGT1 in complex with full GSH molecule | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLUTATHIONE, ... | | Authors: | Terzyan, S.S, Hanigan, M. | | Deposit date: | 2020-07-08 | | Release date: | 2020-11-25 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structures of glutathione- and inhibitor-bound human GGT1: critical interactions within the cysteinylglycine binding site.
J.Biol.Chem., 296, 2020
|
|
7OJS
 
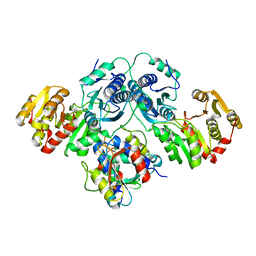 | |
7OJR
 
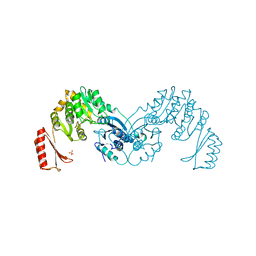 | |
5EYF
 
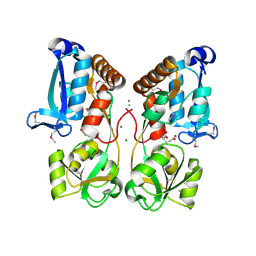 | | Crystal Structure of Solute-binding Protein from Enterococcus faecium with Bound Glutamate | | Descriptor: | CHLORIDE ION, GLUTAMIC ACID, Glutamate ABC superfamily ATP binding cassette transporter, ... | | Authors: | Maltseva, N, Kim, Y, Mulligan, R, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-11-24 | | Release date: | 2015-12-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal Structure of Solute-binding Protein from Enterococcus faecium with Bound Glutamate
To Be Published
|
|
2FE6
 
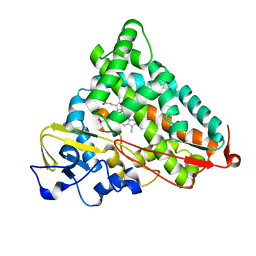 | | P450CAM from Pseudomonas putida reconstituted with manganic protoporphyrin IX | | Descriptor: | Cytochrome P450-cam, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING MN | | Authors: | von Koenig, K, Makris, T.M, Sligar, S.G, Schlichting, I. | | Deposit date: | 2005-12-15 | | Release date: | 2006-03-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The status of high-valent metal oxo complexes in the P450 cytochromes.
J.Inorg.Biochem., 100, 2006
|
|
6Y84
 
 | | SARS-CoV-2 main protease with unliganded active site (2019-nCoV, coronavirus disease 2019, COVID-19) | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Owen, C.D, Lukacik, P, Strain-Damerell, C.M, Douangamath, A, Powell, A.J, Fearon, D, Brandao-Neto, J, Crawshaw, A.D, Aragao, D, Williams, M, Flaig, R, Hall, D.R, McAuley, K.E, Mazzorana, M, Stuart, D.I, von Delft, F, Walsh, M.A. | | Deposit date: | 2020-03-03 | | Release date: | 2020-03-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | COVID-19 main protease with unliganded active site
To Be Published
|
|
8W4U
 
 | | human KCNQ2-CaM in complex with PIP2 and HN37 | | Descriptor: | Calmodulin-1, Potassium voltage-gated channel subfamily KQT member 2, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate, ... | | Authors: | Ma, D, Li, X, Guo, J. | | Deposit date: | 2023-08-25 | | Release date: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Ligand activation mechanisms of human KCNQ2 channel.
Nat Commun, 14, 2023
|
|
7U2K
 
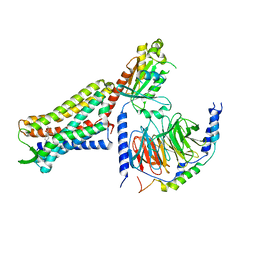 | | C6-guano bound Mu Opioid Receptor-Gi Protein Complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wang, H, Kobilka, B. | | Deposit date: | 2022-02-24 | | Release date: | 2022-12-07 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure-based design of bitopic ligands for the μ-opioid receptor.
Nature, 613, 2023
|
|
2BH5
 
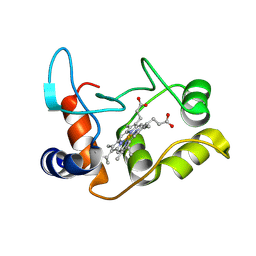 | | X-ray structure of the M100K variant of ferric cyt c-550 from Paracoccus versutus determined at 295 K. | | Descriptor: | CYTOCHROME C-550, HEME C | | Authors: | Worrall, J.A.R, van Roon, A.-M.M, Ubbink, M, Canters, G.W. | | Deposit date: | 2005-01-07 | | Release date: | 2005-05-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Effect of Replacing the Axial Methionine Ligand with a Lysine Residue in Cytochrome C-550 from Paracoccus Versutus Assessed by X-Ray Crystallography and Unfolding.
FEBS J., 272, 2005
|
|
6VJZ
 
 | | CryoEM structure of Hrd1-Usa1/Der1/Hrd3 complex of the expected topology | | Descriptor: | Degradation in the endoplasmic reticulum protein 1, ERAD-associated E3 ubiquitin-protein ligase HRD1, ERAD-associated E3 ubiquitin-protein ligase component HRD3, ... | | Authors: | Wu, X, Rapoport, T.A. | | Deposit date: | 2020-01-18 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis of ER-associated protein degradation mediated by the Hrd1 ubiquitin ligase complex.
Science, 368, 2020
|
|
7U0U
 
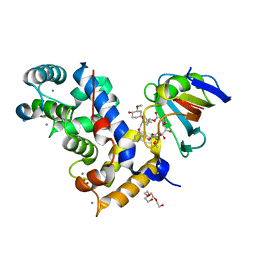 | | Crystal Structure of a Aspergillus fumigatus Calcineurin A - Calcineurin B fusion bound to FKBP12 and FK-506 | | Descriptor: | 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, CALCIUM ION, PHOSPHATE ION, ... | | Authors: | Fox III, D, Abendroth, J, DeBouver, N.D, Hoy, M.J, Heitman, J, Lorimer, D.D, Horanyi, P.S, Edwards, T.E, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-02-18 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Guided Synthesis of FK506 and FK520 Analogs with Increased Selectivity Exhibit In Vivo Therapeutic Efficacy against Cryptococcus.
Mbio, 13, 2022
|
|
7OVO
 
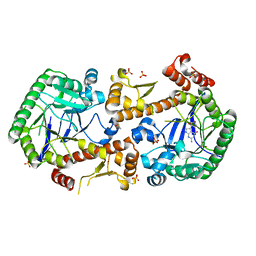 | | Heterodimeric murine tRNA-guanine transglycosylase in complex with queuine | | Descriptor: | 2-amino-5-({[(1S,4S,5R)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, DIMETHYL SULFOXIDE, Queuine tRNA-ribosyltransferase accessory subunit 2, ... | | Authors: | Sebastiani, M, Heine, A, Reuter, K. | | Deposit date: | 2021-06-15 | | Release date: | 2022-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Biochemical Investigation of the Heterodimeric Murine tRNA-Guanine Transglycosylase.
Acs Chem.Biol., 17, 2022
|
|
7OV9
 
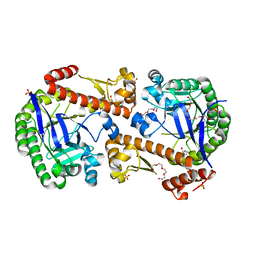 | | Heterodimeric tRNA-Guanine Transglycosylase from mouse, apo-structure | | Descriptor: | HEXAETHYLENE GLYCOL, Queuine tRNA-ribosyltransferase accessory subunit 2, Queuine tRNA-ribosyltransferase catalytic subunit 1, ... | | Authors: | Sebastiani, M, Heine, A, Reuter, K. | | Deposit date: | 2021-06-14 | | Release date: | 2022-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Biochemical Investigation of the Heterodimeric Murine tRNA-Guanine Transglycosylase.
Acs Chem.Biol., 17, 2022
|
|
7OVS
 
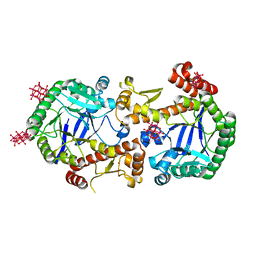 | | Heterodimeric murine tRNA-guanine transglycosylase in the presence of Anderson-Evans type (TEW) and Strandberg type polyoxometalate (POM) | | Descriptor: | 6-tungstotellurate(VI), Queuine tRNA-ribosyltransferase accessory subunit 2, Queuine tRNA-ribosyltransferase catalytic subunit 1, ... | | Authors: | Sebastiani, M, Heine, A, Reuter, K. | | Deposit date: | 2021-06-15 | | Release date: | 2022-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Biochemical Investigation of the Heterodimeric Murine tRNA-Guanine Transglycosylase.
Acs Chem.Biol., 17, 2022
|
|
6VK1
 
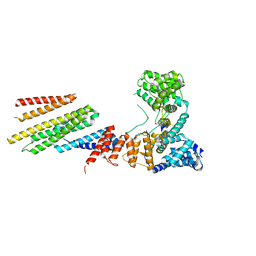 | | CryoEM structure of Hrd1/Hrd3 part from Hrd1-Usa1/Der1/Hrd3 complex | | Descriptor: | ERAD-associated E3 ubiquitin-protein ligase HRD1, ERAD-associated E3 ubiquitin-protein ligase component HRD3 | | Authors: | Wu, X, Rapoport, T.A. | | Deposit date: | 2020-01-18 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of ER-associated protein degradation mediated by the Hrd1 ubiquitin ligase complex.
Science, 368, 2020
|
|
