7ZAC
 
 | | Sam68 | | Descriptor: | 1,2-ETHANEDIOL, KHDR1 protein, NITRATE ION | | Authors: | Nadal, M, Fuentes-Prior, P. | | Deposit date: | 2022-03-22 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structure and function analysis of Sam68 and hnRNP A1 synergy in the exclusion of exon 7 from SMN2 transcripts.
Protein Sci., 32, 2023
|
|
7ZAF
 
 | | Sam68 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Nadal, M, Puestes-Prior, P. | | Deposit date: | 2022-03-22 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structure and function analysis of Sam68 and hnRNP A1 synergy in the exclusion of exon 7 from SMN2 transcripts.
Protein Sci., 32, 2023
|
|
7ZAM
 
 | | Sam68 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, IODIDE ION, ... | | Authors: | Nadal, M, Fuentes-Prior, P. | | Deposit date: | 2022-03-22 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structure and function analysis of Sam68 and hnRNP A1 synergy in the exclusion of exon 7 from SMN2 transcripts.
Protein Sci., 32, 2023
|
|
3GXP
 
 | | Crystal structure of acid-alpha-galactosidase A complexed with galactose at pH 4.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-galactosidase A, ... | | Authors: | Lieberman, R.L. | | Deposit date: | 2009-04-02 | | Release date: | 2009-05-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Effects of pH and iminosugar pharmacological chaperones on lysosomal glycosidase structure and stability.
Biochemistry, 48, 2009
|
|
7ZJW
 
 | | Rabbit 80S ribosome as it decodes the Sec-UGA codon | | Descriptor: | 18S rRNA, 28S rRNA, 40S Ribosomal protein eS19, ... | | Authors: | Hilal, T, Simonovic, M, Spahn, C.M.T. | | Deposit date: | 2022-04-12 | | Release date: | 2022-10-19 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure of the mammalian ribosome as it decodes the selenocysteine UGA codon.
Science, 376, 2022
|
|
3GXF
 
 | |
4B88
 
 | |
3GXT
 
 | |
7ZSI
 
 | | Structure of Orange Carotenoid Protein with canthaxanthin bound after 5 minutes of illumination | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Chukhutsina, V.U, Baxter, J.M, Fadini, A, Morgan, R.M, Pope, M.A, Maghlaoui, K, Orr, C, Wagner, A, van Thor, J.J. | | Deposit date: | 2022-05-06 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Light activation of Orange Carotenoid Protein reveals bicycle-pedal single-bond isomerization.
Nat Commun, 13, 2022
|
|
7ZSH
 
 | | Structure of Orange Carotenoid Protein with canthaxanthin bound after 2 minutes of illumination | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Chukhutsina, V.U, Baxter, J.M, Fadini, A, Morgan, R.M, Pope, M.A, Maghlaoui, K, Orr, C, Wagner, A, van Thor, J.J. | | Deposit date: | 2022-05-06 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Light activation of Orange Carotenoid Protein reveals bicycle-pedal single-bond isomerization.
Nat Commun, 13, 2022
|
|
7ZSF
 
 | | Structure of Orange Carotenoid Protein with canthaxanthin bound | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Chukhutsina, V.U, Baxter, J.M, Fadini, A, Morgan, R.M, Pope, M.A, Maghlaoui, K, Orr, C, Wagner, A, van Thor, J.J. | | Deposit date: | 2022-05-06 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Light activation of Orange Carotenoid Protein reveals bicycle-pedal single-bond isomerization.
Nat Commun, 13, 2022
|
|
7ZSJ
 
 | | Structure of Orange Carotenoid Protein with canthaxanthin bound after 10 minutes of illumination | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Chukhutsina, V.U, Baxter, J.M, Fadini, A, Morgan, R.M, Pope, M.A, Maghlaoui, K, Orr, C, Wagner, A, van Thor, J.J. | | Deposit date: | 2022-05-06 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Light activation of Orange Carotenoid Protein reveals bicycle-pedal single-bond isomerization.
Nat Commun, 13, 2022
|
|
7ZQ0
 
 | | Room temperature SSX structure of GH11 xylanase from Nectria haematococca (1000 frames) | | Descriptor: | Endo-1,4-beta-xylanase | | Authors: | Oberthuer, D, Andaleeb, H, Betzel, C, Perbandt, M, Yefanov, O, Zielinski, K. | | Deposit date: | 2022-04-29 | | Release date: | 2022-11-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rapid and efficient room-temperature serial synchrotron crystallography using the CFEL TapeDrive.
Iucrj, 9, 2022
|
|
7ZSG
 
 | | Structure of Orange Carotenoid Protein with canthaxanthin bound after 1 minute of illumination | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Chukhutsina, V.U, Baxter, J.M, Fadini, A, Morgan, R.M, Pope, M.A, Maghlaoui, K, Orr, C, Wagner, A, van Thor, J.J. | | Deposit date: | 2022-05-06 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Light activation of Orange Carotenoid Protein reveals bicycle-pedal single-bond isomerization.
Nat Commun, 13, 2022
|
|
7ZPV
 
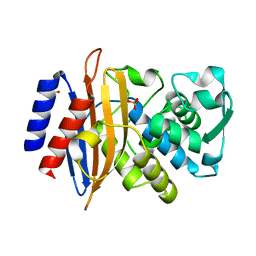 | | Room temperature SSX crystal structure of CTX-M-14 | | Descriptor: | Beta-lactamase, SULFATE ION | | Authors: | Oberthuer, D, Perbandt, M, Prester, A, Rohde, H, Betzel, C, Yefanov, O. | | Deposit date: | 2022-04-29 | | Release date: | 2022-11-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Rapid and efficient room-temperature serial synchrotron crystallography using the CFEL TapeDrive.
Iucrj, 9, 2022
|
|
2Z12
 
 | |
1M5D
 
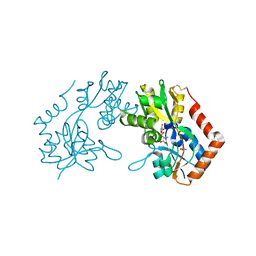 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J-Y702F) IN COMPLEX WITH Br-HIBO AT 1.73 A RESOLUTION | | Descriptor: | (S)-2-AMINO-3-(4-BROMO-3-HYDROXY-ISOXAZOL-5-YL)PROPIONIC ACID, Glutamate receptor 2, SULFATE ION | | Authors: | Hogner, A, Kastrup, J.S, Jin, R, Liljefors, T, Mayer, M.L, Egebjerg, J, Larsen, I.K, Gouaux, E. | | Deposit date: | 2002-07-09 | | Release date: | 2002-09-18 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural Basis for AMPA Receptor Activation and Ligand Selectivity:
Crystal Structures of Five Agonist Complexes with the GluR2 Ligand-binding
Core
J.Mol.Biol., 322, 2002
|
|
3DJG
 
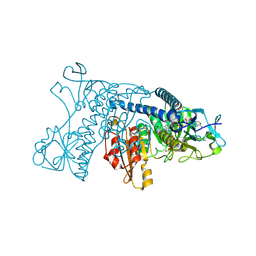 | | Catalytic cycle of human glutathione reductase near 1 A resolution | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Glutathione reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Berkholz, D.S, Faber, H.R, Savvides, S.N, Karplus, P.A. | | Deposit date: | 2008-06-23 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Catalytic cycle of human glutathione reductase near 1 A resolution.
J.Mol.Biol., 382, 2008
|
|
1M5F
 
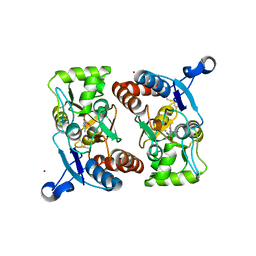 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J-Y702F) IN COMPLEX WITH ACPA AT 1.95 A RESOLUTION | | Descriptor: | (S)-2-AMINO-3-(3-CARBOXY-5-METHYLISOXAZOL-4-YL)PROPIONIC ACID, ACETATE ION, Glutamate receptor 2, ... | | Authors: | Hogner, A, Kastrup, J.S, Jin, R, Liljefors, T, Mayer, M.L, Egebjerg, J, Larsen, I.K, Gouaux, E. | | Deposit date: | 2002-07-09 | | Release date: | 2002-09-18 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Basis for AMPA Receptor Activation and Ligand Selectivity:
Crystal Structures of Five Agonist Complexes with the GluR2 Ligand-binding
Core
J.Mol.Biol., 322, 2002
|
|
1S8G
 
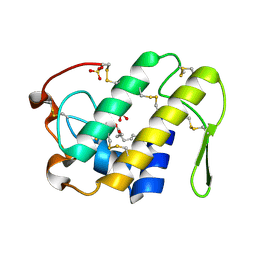 | | Crystal structure of Lys49-Phospholipase A2 from Agkistrodon contortrix laticinctus, fatty acid bound form | | Descriptor: | GLYCEROL, LAURIC ACID, Phospholipase A2 homolog, ... | | Authors: | Ambrosio, A.L.B, de Souza, D.H.F, Nonato, M.C, Selistre de Araujo, H.S, Ownby, C.L, Garratt, R.C. | | Deposit date: | 2004-02-02 | | Release date: | 2004-02-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Molecular Mechanism for Lys49-Phospholipase A2 Activity Based on Ligand-induced Conformational Change.
J.Biol.Chem., 280, 2005
|
|
3DMG
 
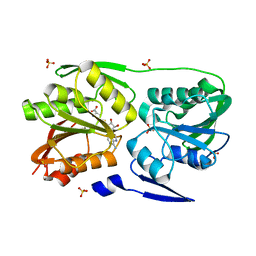 | | T. Thermophilus 16S rRNA N2 G1207 methyltransferase (RsmC) in complex with AdoHcy | | Descriptor: | Probable ribosomal RNA small subunit methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION | | Authors: | Demirci, H, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2008-07-01 | | Release date: | 2008-07-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of the Thermus thermophilus 16 S rRNA Methyltransferase RsmC in Complex with Cofactor and Substrate Guanosine.
J.Biol.Chem., 283, 2008
|
|
3D9A
 
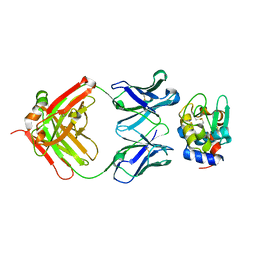 | | High Resolution Crystal Structure Structure of HyHel10 Fab Complexed to Hen Egg Lysozyme | | Descriptor: | Heavy Chain of HyHel10 Antibody Fragment (Fab), Light Chain of HyHel10 Antibody Fragment (Fab), Lysozyme C | | Authors: | DeSantis, M.E, Li, M, Shanmuganathan, A, Acchione, M, Walter, R, Wlodawer, A, Smith-Gill, S. | | Deposit date: | 2008-05-27 | | Release date: | 2008-06-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Light chain somatic mutations change thermodynamics of binding and water coordination in the HyHEL-10 family of antibodies.
Mol.Immunol., 47, 2009
|
|
4ARJ
 
 | | Crystal structure of a pesticin (translocation and receptor binding domain) from Y. pestis and T4-lysozyme chimera | | Descriptor: | PESTICIN, LYSOZYME, SULFATE ION | | Authors: | Zeth, K, Patzer, S.I, Albrecht, R, Braun, V. | | Deposit date: | 2012-04-24 | | Release date: | 2012-05-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.593 Å) | | Cite: | Structure and Mechanistic Studies of Pesticin, a Bacterial Homolog of Phage Lysozymes.
J.Biol.Chem., 287, 2012
|
|
3GXM
 
 | |
3GXD
 
 | | Crystal structure of Apo acid-beta-glucosidase pH 4.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glucosylceramidase, PHOSPHATE ION | | Authors: | Lieberman, R.L. | | Deposit date: | 2009-04-02 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Effects of pH and iminosugar pharmacological chaperones on lysosomal glycosidase structure and stability.
Biochemistry, 48, 2009
|
|
