8V9I
 
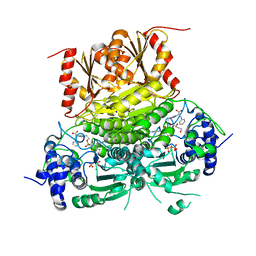 | | 1-deoxy-D-xylulose 5-phosphate synthase (DXPS) from Deinococcus radiodurans with D-phenylalanine-derived triazole acetylphosphonate (D-PheTrAP) bound | | Descriptor: | 1-deoxy-D-xylulose-5-phosphate synthase, 2-HYDROXY BUTANE-1,4-DIOL, 3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-2-{(1S)-1-[(S)-(2-{1-[(1R)-1-carboxy-2-phenylethyl]-1H-1,2,3-triazol-4-yl}ethoxy)(hydroxy)phosphoryl]-1-hydroxyethyl}-5-(2-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3-thiazol-3-ium, ... | | Authors: | Chen, P.Y.-T, Drennan, C.L. | | Deposit date: | 2023-12-08 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Potent Inhibition of E. coli DXP Synthase by a gem -Diaryl Bisubstrate Analog.
Acs Infect Dis., 10, 2024
|
|
8XC4
 
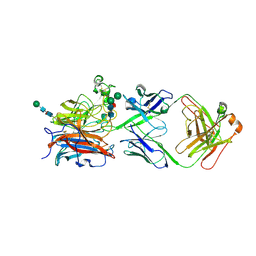 | | Nipah virus attachment glycoprotein head domain in complex with a broadly neutralizing antibody 1E5 | | Descriptor: | 1E5-VH, 1E5-VL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fan, P.F, Yu, C.M, Chen, W. | | Deposit date: | 2023-12-08 | | Release date: | 2024-01-24 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | A potent Henipavirus cross-neutralizing antibody reveals a dynamic fusion-triggering pattern of the G-tetramer.
Nat Commun, 15, 2024
|
|
8V9K
 
 | | Cryo-EM structure of the Mycobacterium smegmatis 70S ribosome in complex with hibernation factor Rv2629 (Balon) (Structure 5) | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S2, ... | | Authors: | Rybak, M.Y, Helena-Bueno, K, Hill, C.H, Melnikov, S.V, Gagnon, M.G. | | Deposit date: | 2023-12-08 | | Release date: | 2024-02-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A new family of bacterial ribosome hibernation factors.
Nature, 626, 2024
|
|
8V9L
 
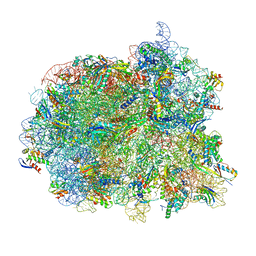 | | Cryo-EM structure of the Mycobacterium smegmatis 70S ribosome in complex with hibernation factor Msmeg1130 (Balon) and MsmegEF-Tu(GDP) (Composite structure 6) | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S2, ... | | Authors: | Rybak, M.Y, Helena-Bueno, K, Hill, C.H, Melnikov, S.V, Gagnon, M.G. | | Deposit date: | 2023-12-08 | | Release date: | 2024-02-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A new family of bacterial ribosome hibernation factors.
Nature, 626, 2024
|
|
8RD8
 
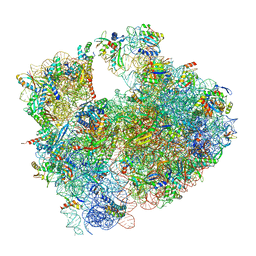 | | Cryo-EM structure of P. urativorans 70S ribosome in complex with hibernation factors Balon and RaiA (structure 1). | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S30, ... | | Authors: | Helena-Bueno, K, Rybak, M.Y, Gagnon, M.G, Hill, C.H, Melnikov, S.V. | | Deposit date: | 2023-12-07 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | A new family of bacterial ribosome hibernation factors.
Nature, 626, 2024
|
|
8RDA
 
 | |
8RCS
 
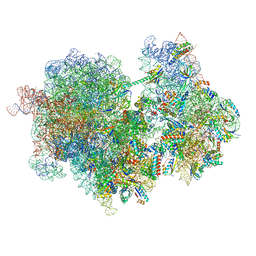 | |
8RCT
 
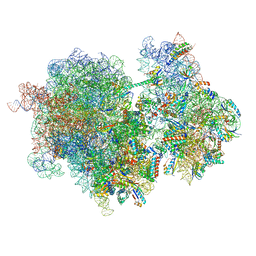 | |
8RD3
 
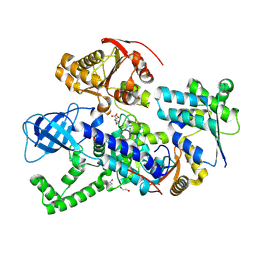 | |
8RCV
 
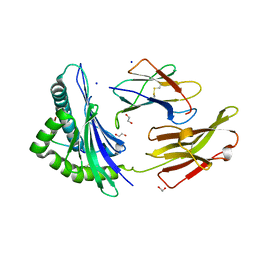 | | Crystal structure of HLA B*13:01 in complex with SVLNDIFSRL, an 10-mer epitope from SARS-CoV-2 Spike (S975-984) | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, HLA class I histocompatibility antigen B alpha chain, ... | | Authors: | Ahn, Y.M, Maddumage, J.C, Szeto, C, Gras, S. | | Deposit date: | 2023-12-07 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The impact of SARS-CoV-2 spike mutation on peptide presentation is HLA allomorph-specific.
Curr Res Struct Biol, 7, 2024
|
|
8XBW
 
 | | The cryo-EM structure of the RAD51 N-terminal lobe domain bound to the histone H4 tail of the nucleosome | | Descriptor: | DNA (5'-D(P*AP*CP*CP*GP*CP*TP*TP*AP*AP*AP*CP*GP*CP*AP*CP*GP*TP*A)-3'), DNA (5'-D(P*TP*AP*CP*GP*TP*GP*CP*GP*TP*TP*TP*AP*AP*GP*CP*GP*GP*T)-3'), DNA repair protein RAD51 homolog 1, ... | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-12-07 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBU
 
 | | The cryo-EM structure of the decameric RAD51 ring bound to the nucleosome with the linker DNA binding | | Descriptor: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-12-07 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.24 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBT
 
 | | The cryo-EM structure of the octameric RAD51 ring bound to the nucleosome with the linker DNA binding | | Descriptor: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-12-07 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.12 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XC1
 
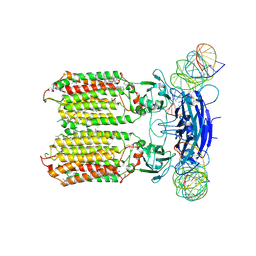 | | C. elegans SID1 in complex with dsRNA | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gong, D.S. | | Deposit date: | 2023-12-07 | | Release date: | 2024-06-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.21 Å) | | Cite: | Structural basis for double-stranded RNA recognition by SID1.
Nucleic Acids Res., 52, 2024
|
|
8XBS
 
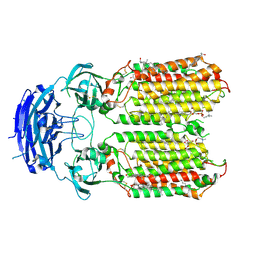 | | C. elegans apo-SID1 structure | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gong, D.S. | | Deposit date: | 2023-12-07 | | Release date: | 2024-06-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.21 Å) | | Cite: | Structural basis for double-stranded RNA recognition by SID1.
Nucleic Acids Res., 52, 2024
|
|
8RCQ
 
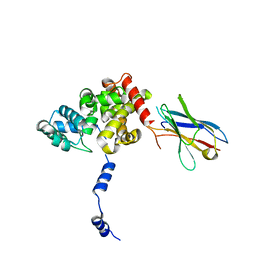 | | Structural flexibility of Nucleoprotein of the Toscana virus in the presence of a nanobody. | | Descriptor: | Nucleoprotein, VHH | | Authors: | Papageorgiou, N, Ferron, F, Coutard, B, Lichiere, J, Baklouti, A. | | Deposit date: | 2023-12-06 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural flexibility of Toscana virus nucleoprotein in the presence of a single-chain camelid antibody.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8RC4
 
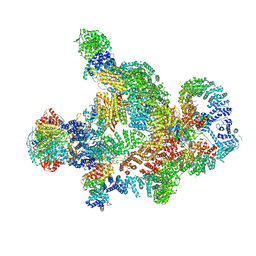 | | Structure of Integrator-PP2A complex | | Descriptor: | DSS1, Integrator complex subunit 1, Integrator complex subunit 10, ... | | Authors: | Fianu, I, Ochmann, M, Walshe, J.L, Cramer, P. | | Deposit date: | 2023-12-06 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of Integrator-dependent RNA polymerase II termination.
Nature, 629, 2024
|
|
8RCO
 
 | | Structure of Human Serum Albumin in complex with Aristolochic Acid II at 1.9 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 6-nitronaphtho[1,2-e][1,3]benzodioxole-5-carboxylic acid, MYRISTIC ACID, ... | | Authors: | Pomyalov, S, Sidorenko, V.S, Grollman, A.P, Shoham, G. | | Deposit date: | 2023-12-06 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and mechanistic insights into the transport of aristolochic acids and their active metabolites by human serum albumin.
J.Biol.Chem., 300, 2024
|
|
8RCP
 
 | | Structure of Human Serum Albumin in complex with Myristic Acid | | Descriptor: | 1,2-ETHANEDIOL, MYRISTIC ACID, Serum albumin | | Authors: | Pomyalov, S, Sidorenko, V.S, Grollman, A.P, Shoham, G. | | Deposit date: | 2023-12-06 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and mechanistic insights into the transport of aristolochic acids and their active metabolites by human serum albumin.
J.Biol.Chem., 300, 2024
|
|
8RCM
 
 | |
8RCL
 
 | |
8RBX
 
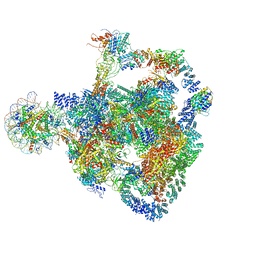 | | Structure of Integrator-PP2A bound to a paused RNA polymerase II-DSIF-NELF-nucleosome complex | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB11-a, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Fianu, I, Ochmann, M, Walshe, J.L, Cramer, P. | | Deposit date: | 2023-12-05 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of Integrator-dependent RNA polymerase II termination.
Nature, 629, 2024
|
|
8RBZ
 
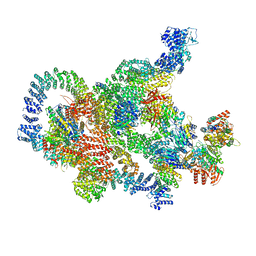 | | Structure of Integrator-PP2A-SOSS-CTD post-termination complex | | Descriptor: | DNA-directed RNA polymerase subunit, DSS1, Integrator complex subunit 1, ... | | Authors: | Fianu, I, Ochmann, M, Walshe, J.L, Cramer, P. | | Deposit date: | 2023-12-05 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of Integrator-dependent RNA polymerase II termination.
Nature, 629, 2024
|
|
8RC0
 
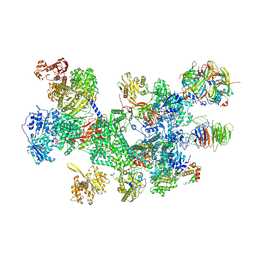 | | Structure of the human 20S U5 snRNP | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, CD2 antigen cytoplasmic tail-binding protein 2, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Schneider, S, Galej, W.P. | | Deposit date: | 2023-12-05 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the human 20S U5 snRNP.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8RC1
 
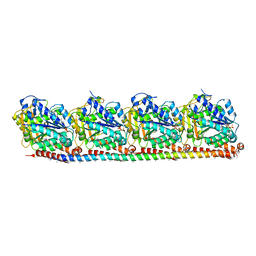 | |
