1JUX
 
 | |
1D7S
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX OF 2,2-DIALKYLGLYCINE DECARBOXYLASE WITH DCS | | Descriptor: | D-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-N,O-CYCLOSERYLAMIDE, POTASSIUM ION, PROTEIN (2,2-DIALKYLGLYCINE DECARBOXYLASE (PYRUVATE)), ... | | Authors: | Malashkevich, V.N, Toney, M.D, Strop, P, Keller, J, Jansonius, J.N. | | Deposit date: | 1999-10-19 | | Release date: | 1999-11-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of dialkylglycine decarboxylase inhibitor complexes.
J.Mol.Biol., 294, 1999
|
|
5NH7
 
 | |
3N4N
 
 | | Insights into the stabilizing contributions of a bicyclic cytosine analogue: crystal structures of DNA duplexes containing 7,8-dihydropyrido[2,3-d]pyrimidin-2-one | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*A)-3', 5'-D(P*TP*TP*(B7C)P*GP*CP*G)-3' | | Authors: | Takenaka, A, Juan, E.C.M, Shimizu, S. | | Deposit date: | 2010-05-22 | | Release date: | 2010-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Insights into the DNA stabilizing contributions of a bicyclic cytosine analogue: crystal structures of DNA duplexes containing 7,8-dihydropyrido [2,3-d]pyrimidin-2-one.
Nucleic Acids Res., 2010
|
|
2YMZ
 
 | | Crystal structure of chicken Galectin 2 | | Descriptor: | GALECTIN 2, SULFATE ION, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Fernandez, I.S, Ruiz, F.M, Solis, D, Gabius, H.-J, Romero, A. | | Deposit date: | 2012-10-11 | | Release date: | 2013-08-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Fine-Tuning of Prototype Chicken Galectins: Structure of Cg-2 and Structure-Activity Correlations
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1FQP
 
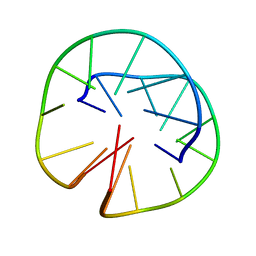 | | INTRAMOLECULAR QUADRUPLEX DNA WITH THREE GGGG REPEATS, NMR, PH 6.7, 0.1 M NA+ AND 4 MM (STRAND CONCENTRATION), 5 STRUCTURES | | Descriptor: | DNA (5'-D(GP*GP*GP*TP*TP*TP*TP*GP*GP*G)-3') | | Authors: | Keniry, M.A, Strahan, G.D, Owen, E.A, Shafer, R.H. | | Deposit date: | 1996-08-01 | | Release date: | 1997-02-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Na+ form of the dimeric guanine quadruplex [d(G3T4G3)]2.
Eur.J.Biochem., 233, 1995
|
|
4KT7
 
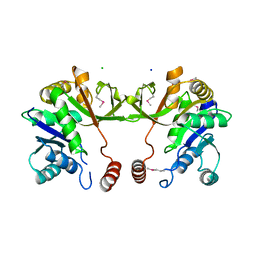 | | The crystal structure of 4-diphosphocytidyl-2C-methyl-D-erythritolsynthase from Anaerococcus prevotii DSM 20548 | | Descriptor: | 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, CHLORIDE ION, SODIUM ION | | Authors: | Borek, D, Tan, K, Stols, L, Eschenfeidt, W.H, Otwinoski, Z, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-20 | | Release date: | 2013-06-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | The crystal structure of 4-diphosphocytidyl-2C-methyl-D-erythritolsynthase from Anaerococcus prevotii DSM 20548
To be Published
|
|
1XME
 
 | | Structure of Recombinant Cytochrome ba3 Oxidase from Thermus thermophilus | | Descriptor: | COPPER (II) ION, Cytochrome c oxidase polypeptide I, Cytochrome c oxidase polypeptide II, ... | | Authors: | Hunsicker-Wang, L.M, Pacoma, R.L, Chen, Y, Fee, J.A, Stout, C.D. | | Deposit date: | 2004-10-01 | | Release date: | 2005-03-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A novel cryoprotection scheme for enhancing the diffraction of crystals of recombinant cytochrome ba3 oxidase from Thermus thermophilus.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
4CBO
 
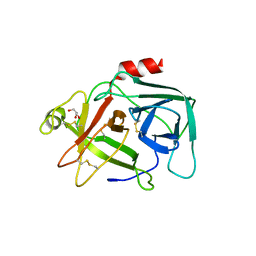 | |
245D
 
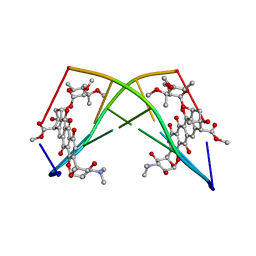 | | DNA-DRUG REFINEMENT: A COMPARISON OF THE PROGRAMS NUCLSQ, PROLSQ, SHELXL93 AND X-PLOR, USING THE LOW TEMPERATURE D(TGATCA)-NOGALAMYCIN STRUCTURE | | Descriptor: | DNA (5'-D(*TP*GP*AP*TP*CP*A)-3'), NOGALAMYCIN | | Authors: | Schuerman, G.S, Smith, C.K, Turkenburg, J.P, Dettmar, A.N, Van Meervelt, L, Moore, M.H. | | Deposit date: | 1996-01-12 | | Release date: | 1996-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | DNA-drug refinement: a comparison of the programs NUCLSQ, PROLSQ, SHELXL93 and X-PLOR, using the low-temperature d(TGATCA)-nogalamycin structure.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
4CST
 
 | | Crystal structure of FimH in complex with 3'-Chloro-4'-(alpha-D-mannopyranosyloxy)-biphenyl-4-carbonitrile | | Descriptor: | 3'-chloro-4'-(alpha-D-mannopyranosyloxy)biphenyl-4-carbonitrile, PROTEIN FIMH | | Authors: | Kleeb, S, Pang, L, Mayer, K, Sigl, A, Eris, D, Preston, R.C, Zihlmann, P, Abgottspon, D, Hutter, A, Scharenberg, M, Jian, X, Navarra, G, Rabbani, S, Smiesko, M, Luedin, N, Jakob, R.P, Schwardt, O, Maier, T, Sharpe, T, Ernst, B. | | Deposit date: | 2014-03-10 | | Release date: | 2015-02-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Fimh Antagonists: Bioisosteres to Improve the in Vitro and in Vivo Pk/Pd Profile.
J.Med.Chem., 58, 2015
|
|
1D82
 
 | |
224D
 
 | | DNA-DRUG REFINEMENT: A COMPARISON OF THE PROGRAMS NUCLSQ, PROLSQ, SHELXL93 AND X-PLOR, USING THE LOW TEMPERATURE D(TGATCA)-NOGALAMYCIN STRUCTURE | | Descriptor: | DNA (5'-D(*TP*GP*AP*TP*CP*A)-3'), NOGALAMYCIN | | Authors: | Schuerman, G.S, Smith, C.K, Turkenburg, J.P, Dettmar, A.N, Van Meervelt, L, Moore, M.H. | | Deposit date: | 1995-08-01 | | Release date: | 1995-11-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | DNA-drug refinement: a comparison of the programs NUCLSQ, PROLSQ, SHELXL93 and X-PLOR, using the low-temperature d(TGATCA)-nogalamycin structure.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
5E5W
 
 | | Hemagglutinin-esterase-fusion mutant structure of influenza D virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin-esterase, ... | | Authors: | Song, H, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2015-10-09 | | Release date: | 2016-03-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An Open Receptor-Binding Cavity of Hemagglutinin-Esterase-Fusion Glycoprotein from Newly-Identified Influenza D Virus: Basis for Its Broad Cell Tropism
PLoS Pathog., 12, 2016
|
|
1V41
 
 | | Crystal structure of human PNP complexed with 8-Azaguanine | | Descriptor: | 5-AMINO-1H-[1,2,3]TRIAZOLO[4,5-D]PYRIMIDIN-7-OL, Purine nucleoside phosphorylase, SULFATE ION | | Authors: | Dos Santos, D.M, Canduri, F, Silva, R.G, Mendes, M.A, Basso, L.A, Palma, M.S, De Azevedo Jr, W.F, Santos, D.S. | | Deposit date: | 2003-11-08 | | Release date: | 2004-12-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of human PNP complexed with ligands.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
5E64
 
 | | Hemagglutinin-esterase-fusion protein structure of influenza D virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CACODYLATE ION, ... | | Authors: | Song, H, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2015-10-09 | | Release date: | 2016-03-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An Open Receptor-Binding Cavity of Hemagglutinin-Esterase-Fusion Glycoprotein from Newly-Identified Influenza D Virus: Basis for Its Broad Cell Tropism
PLoS Pathog., 12, 2016
|
|
4IED
 
 | | Crystal Structure of FUS-1 (OXA-85), a Class D beta-lactamase from Fusobacterium nucleatum subsp. polymorphum | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Class D beta-lactamase, ... | | Authors: | Mangani, S, Benvenuti, M, Docquier, J.D. | | Deposit date: | 2012-12-13 | | Release date: | 2014-01-22 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of FUS-1 (OXA-85), a Class D beta-lactamase from Fusobacterium nucleatum subsp. polymorphum
To be Published
|
|
3L8E
 
 | | Crystal Structure of apo form of D,D-heptose 1.7-bisphosphate phosphatase from E. Coli | | Descriptor: | ACETIC ACID, D,D-heptose 1,7-bisphosphate phosphatase, ZINC ION | | Authors: | Nguyen, H, Peisach, E, Allen, K.N. | | Deposit date: | 2009-12-31 | | Release date: | 2010-02-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural Determinants of Substrate Recognition in the HAD Superfamily Member d-glycero-d-manno-Heptose-1,7-bisphosphate Phosphatase (GmhB) .
Biochemistry, 49, 2010
|
|
1E3U
 
 | | MAD structure of OXA10 class D beta-lactamase | | Descriptor: | 1,2-ETHANEDIOL, BETA-LACTAMASE OXA-10, GOLD (I) CYANIDE ION, ... | | Authors: | Maveyraud, L, Golemi, D, Kotra, L.P, Tranier, S, Vakulenko, S, Mobashery, S, Samama, J.P. | | Deposit date: | 2000-06-23 | | Release date: | 2001-01-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Insights Into Class D Beta-Lactamases are Revealed by the Crystal Structure of the Oxa10 Enzyme from Pseudomonas Aeruginosa
Structure, 8, 2000
|
|
4MIJ
 
 | | Crystal structure of a Trap periplasmic solute binding protein from Polaromonas sp. JS666 (Bpro_3107), target EFI-510173, with bound alpha/beta D-Galacturonate, space group P21 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, TRAP dicarboxylate transporter, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Zhao, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-08-31 | | Release date: | 2013-09-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4A6S
 
 | | Structure of the PAIL lectin from Pseudomonas aeruginosa in complex with 2-Naphtyl-1-thio-beta-D-galactopyranoside | | Descriptor: | CALCIUM ION, PA-I GALACTOPHILIC LECTIN, naphthalen-2-yl 1-thio-beta-D-galactopyranoside | | Authors: | Rodrigue, J, Ganne, G, Blanchard, B, Saucier, C, Giguere, D, Chiao, T.S, Varrot, A, Imberty, A, Roy, R. | | Deposit date: | 2011-11-08 | | Release date: | 2012-10-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Aromatic Thioglycoside Inhibitors Against the Virulence Factor Leca from Pseudomonas Aeruginosa.
Org.Biomol.Chem., 11, 2013
|
|
4N91
 
 | | Crystal structure of a trap periplasmic solute binding protein from anaerococcus prevotii dsm 20548 (Apre_1383), target EFI-510023, with bound alpha/beta d-glucuronate | | Descriptor: | CHLORIDE ION, TETRAETHYLENE GLYCOL, TRAP dicarboxylate transporter, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-18 | | Release date: | 2013-11-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4DDA
 
 | | EVAL processed HEWL, NAG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lysozyme C | | Authors: | Tanley, S.W, Schreurs, A.M, Kroon-Batenburg, L.M, Meredith, J, Prendergast, R, Walsh, D, Bryant, P, Levy, C, Helliwell, J.R. | | Deposit date: | 2012-01-18 | | Release date: | 2012-04-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural studies of the effect that dimethyl sulfoxide (DMSO) has on cisplatin and carboplatin binding to histidine in a protein.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1MA9
 
 | | Crystal structure of the complex of human vitamin D binding protein and rabbit muscle actin | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, Alpha Skeletal Muscle, ... | | Authors: | Verboven, C, Bogaerts, I, Waelkens, E, Rabijns, A, Van Baelen, H, Bouillon, R, De Ranter, C. | | Deposit date: | 2002-08-02 | | Release date: | 2003-02-04 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Actin-DBP: the perfect structural fit?
Acta Crystallogr.,Sect.D, 59, 2003
|
|
4NX1
 
 | | Crystal structure of a trap periplasmic solute binding protein from Sulfitobacter sp. nas-14.1, target EFI-510292, with bound alpha-D-taluronate | | Descriptor: | C4-dicarboxylate transport system substrate-binding protein, alpha-D-talopyranuronic acid | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-12-08 | | Release date: | 2014-01-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
