3MUZ
 
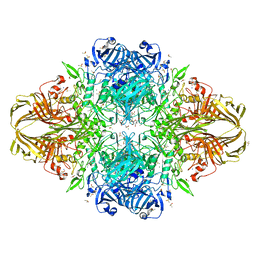 | | E.Coli (lacZ) beta-galactosidase (R599A) in complex with IPTG | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Dugdale, M.L, Vance, M.L, Driedger, M.R, Nibber, A, Tran, A, Huber, R.E. | | Deposit date: | 2010-05-03 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Importance of Arg-599 of b-galactosidase (Escherichia coli) as an anchor for the open conformations of Phe-601 and the active-site loop
Biochem.Cell Biol., 88, 2010
|
|
1LW0
 
 | | CRYSTAL STRUCTURE OF T215Y MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH NEVIRAPINE | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 REVERSE TRANSCRIPTASE, PHOSPHATE ION | | Authors: | Ren, J, Chamberlain, P.P, Nichols, C.E, Douglas, L, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2002-05-30 | | Release date: | 2002-10-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of Zidovudine- or Lamivudine-resistant human immunodeficiency virus type 1 reverse transcriptases containing mutations at codons 41, 184, and 215.
J.Virol., 76, 2002
|
|
9GEJ
 
 | | Crystal structure of CREBBP bromodomain in complex with (2R,13S,E)-2-methyl-1,2,3,5,10,11,13,14,20,21,24,25-dodecahydro-19H,23H-16,18-etheno-9,13-methano-7,28-(metheno)[1,4]diazepino[2,3-k]pyrido[1,2-s][1,4]dioxa[7,19]diazacyclodocosine-4,8-dione | | Descriptor: | 2R,13S,E)-2-methyl-1,2,3,5,10,11,13,14,20,21,24,25-dodecahydro-19H,23H-16,18-etheno-9,13-methano-7,28-(metheno)[1,4]diazepino[2,3-k]pyrido[1,2-s][1,4]dioxa[7,19]diazacyclodocosine-4,8-dione, CREBBP, SULFATE ION | | Authors: | Amann, M, Boyd, A, Einsle, O, Guenther, S, Moroglu, M, Conway, S. | | Deposit date: | 2024-08-07 | | Release date: | 2025-08-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of CREBBP bromodomain in complex with (2R,13S,E)-2-methyl-1,2,3,5,10,11,13,14,20,21,24,25-dodecahydro-19H,23H-16,18-etheno-9,13-methano-7,28-(metheno)[1,4]diazepino[2,3-k]pyrido[1,2-s][1,4]dioxa[7,19]diazacyclodocosine-4,8-dione
To Be Published
|
|
5EXN
 
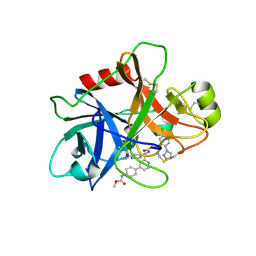 | | FACTOR XIA (C500S [C122S]) IN COMPLEX WITH THE INHIBITOR methyl ~{N}-[4-[2-[(1~{S})-1-[[(~{E})-3-[5-chloranyl-2-(1,2,3,4-tetrazol-1-yl)phenyl]prop-2-enoyl]amino]-2-phenyl-ethyl]pyridin-4-yl]phenyl]carbamate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Coagulation factor XIa light chain, methyl ~{N}-[4-[2-[(1~{S})-1-[[(~{E})-3-[5-chloranyl-2-(1,2,3,4-tetrazol-1-yl)phenyl]prop-2-enoyl]amino]-2-phenyl-ethyl]pyridin-4-yl]phenyl]carbamate | | Authors: | Sheriff, S. | | Deposit date: | 2015-11-23 | | Release date: | 2016-04-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Orally bioavailable pyridine and pyrimidine-based Factor XIa inhibitors: Discovery of the methyl N-phenyl carbamate P2 prime group
Bioorg.Med.Chem., 24, 2016
|
|
2OOL
 
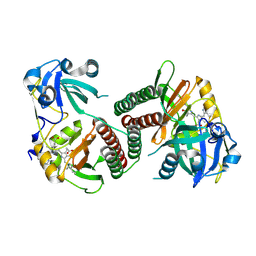 | | Crystal structure of the chromophore-binding domain of an unusual bacteriophytochrome RpBphP3 from R. palustris | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Sensor protein | | Authors: | Yang, X, Stojkovic, E.A, Kuk, J, Moffat, K. | | Deposit date: | 2007-01-25 | | Release date: | 2007-07-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the chromophore binding domain of an unusual bacteriophytochrome, RpBphP3, reveals residues that modulate photoconversion.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
9GEW
 
 | | Crystal structure of CREBBP bromodomain in complex with (R,E)-6-(5-(6-methoxy-2,3-dihydro-4H-benzo[b][1,4]oxazin-4-yl)pent-1-en-1-yl)-4-methyl-1,3,4,5-tetrahydro-2H-benzo[b][1,4]diazepin-2-one | | Descriptor: | (4~{R})-6-[(~{E})-5-(6-methoxy-2,3-dihydro-1,4-benzoxazin-4-yl)pent-1-enyl]-4-methyl-1,3,4,5-tetrahydro-1,5-benzodiazepin-2-one, (R,R)-2,3-BUTANEDIOL, CREBBP | | Authors: | Amann, M, Boyd, A, Einsle, O, Moroglu, M, Conway, S. | | Deposit date: | 2024-08-07 | | Release date: | 2025-08-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystal structure of CREBBP bromodomain in complex with (R,E)-6-(5-(6-methoxy-2,3-dihydro-4H-benzo[b][1,4]oxazin-4-yl)pent-1-en-1-yl)-4-methyl-1,3,4,5-tetrahydro-2H-benzo[b][1,4]diazepin-2-one
To Be Published
|
|
3MV0
 
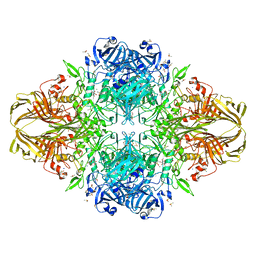 | | E. COLI (lacZ) BETA-GALACTOSIDASE (R599A) IN COMPLEX WITH D-GALCTOPYRANOSYL-1-ONE | | Descriptor: | Beta-galactosidase, D-galactonolactone, DIMETHYL SULFOXIDE, ... | | Authors: | Dugdale, M.L, Vance, M, Driedger, M.L, Nibber, A, Tran, A, Huber, R.E. | | Deposit date: | 2010-05-03 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Importance of Arg-599 of b-galactosidase (Escherichia coli) as an anchor for the open conformations of Phe-601 and the active-site loop
Biochem.Cell Biol., 88, 2010
|
|
9G7A
 
 | | CryoEM structure of human rho1 GABAA receptor in complex with loreclezole | | Descriptor: | 1-[(~{Z})-2-chloranyl-2-(2,4-dichlorophenyl)ethenyl]-1,2,4-triazole, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Fan, C, Howard, R.J, Lindahl, E. | | Deposit date: | 2024-07-20 | | Release date: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (2.12 Å) | | Cite: | CryoEM structure of human rho1 GABAA receptor in complex with neurosteroid
To be published
|
|
9G7B
 
 | | CryoEM structure of human rho1 GABAA receptor in complex with GABA and loreclezole | | Descriptor: | 1-[(~{Z})-2-chloranyl-2-(2,4-dichlorophenyl)ethenyl]-1,2,4-triazole, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Fan, C, Howard, R.J, Lindahl, E. | | Deposit date: | 2024-07-20 | | Release date: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (2.18 Å) | | Cite: | CryoEM structure of human rho1 GABAA receptor in complex with neurosteroid
To be published
|
|
1ZL5
 
 | | Crystal structure of Glu335Gln mutant of Clostridium botulinum neurotoxin E catalytic domain | | Descriptor: | CHLORIDE ION, botulinum neurotoxin type E | | Authors: | Agarwal, R, Binz, T, Swaminathan, S. | | Deposit date: | 2005-05-05 | | Release date: | 2005-07-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Analysis of Active Site Residues of Botulinum Neurotoxin E by Mutational, Functional, and Structural Studies: Glu335Gln Is an Apoenzyme.
Biochemistry, 44, 2005
|
|
6DUG
 
 | | Crystal structure of HIV-1 reverse transcriptase K101P mutant in complex with non-nucleoside inhibitor 25a | | Descriptor: | 1,2-ETHANEDIOL, 4-({4-[(4-{4-[(E)-2-cyanoethenyl]-2,6-dimethylphenoxy}thieno[3,2-d]pyrimidin-2-yl)amino]piperidin-1-yl}methyl)benzene-1-sulfonamide, DIMETHYL SULFOXIDE, ... | | Authors: | Yang, Y, Nguyen, L.A, Smithline, Z.B, Steitz, T.A. | | Deposit date: | 2018-06-20 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.225 Å) | | Cite: | Structural basis for potent and broad inhibition of HIV-1 RT by thiophene[3,2-d]pyrimidine non-nucleoside inhibitors.
Elife, 7, 2018
|
|
9H28
 
 | | Alternative conformation LGTV with TBEV prME | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein E, ... | | Authors: | Bisikalo, K, Rosendal, E. | | Deposit date: | 2024-10-11 | | Release date: | 2024-11-06 | | Last modified: | 2025-10-08 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Influence of the pre-membrane and envelope proteins on structure, pathogenicity, and tropism of tick-borne encephalitis virus.
J.Virol., 99, 2025
|
|
5F6C
 
 | |
8PPZ
 
 | | Co-crystal structure of FKBP12, compound 7 and the FRB fragment of mTOR | | Descriptor: | (1~{S},5~{S},6~{R})-10-[3,5-bis(chloranyl)phenyl]sulfonyl-5-[(~{E})-2-(2-chlorophenyl)ethenyl]-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, ACETATE ION, CALCIUM ION, ... | | Authors: | Meyners, C, Deutscher, R.C.E, Hausch, F. | | Deposit date: | 2023-07-10 | | Release date: | 2023-08-09 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Co-crystal structure of FKBP12, compound 7 and the FRB fragment of mTOR
Chemrxiv, 2023
|
|
1Z9H
 
 | | Microsomal prostaglandin E synthase type-2 | | Descriptor: | ACETATE ION, CHLORIDE ION, INDOMETHACIN, ... | | Authors: | Yamada, T, Komoto, J, Watanabe, K, Ohmiya, Y, Takusagawa, F. | | Deposit date: | 2005-04-02 | | Release date: | 2005-05-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure and Possible Catalytic Mechanism of Microsomal Prostaglandin E Synthase Type 2 (mPGES-2).
J.Mol.Biol., 348, 2005
|
|
3V48
 
 | | Crystal Structure of the putative alpha/beta hydrolase RutD from E.coli | | Descriptor: | GLYCEROL, Putative aminoacrylate hydrolase RutD, THIOCYANATE ION | | Authors: | Knapik, A.A, Petkowski, J.J, Otwinowski, Z, Cymborowski, M.T, Cooper, D.R, Chruszcz, M, Porebski, P.J, Niedzialkowska, E, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-14 | | Release date: | 2012-01-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A multi-faceted analysis of RutD reveals a novel family of alpha / beta hydrolases.
Proteins, 80, 2012
|
|
7A53
 
 | | Structure of DYRK1A in complex with compound 7 | | Descriptor: | (3~{E})-3-[(5-methylfuran-2-yl)methylidene]-1~{H}-indol-2-one, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | Authors: | Dokurno, P, Surgenor, A.E, Hubbard, R.E. | | Deposit date: | 2020-08-20 | | Release date: | 2021-06-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Fragment-Derived Selective Inhibitors of Dual-Specificity Kinases DYRK1A and DYRK1B.
J.Med.Chem., 64, 2021
|
|
6DL7
 
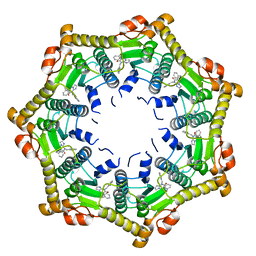 | | Human mitochondrial ClpP in complex with ONC201 (TIC10) | | Descriptor: | 7-benzyl-4-[(2-methylphenyl)methyl]-6,7,8,9-tetrahydroimidazo[1,2-a]pyrido[3,4-e]pyrimidin-5(4H)-one, ATP-dependent Clp protease proteolytic subunit, mitochondrial | | Authors: | Halgas, O, Zarabi, S.F, Schimmer, A, Pai, E.F. | | Deposit date: | 2018-05-31 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mitochondrial ClpP-Mediated Proteolysis Induces Selective Cancer Cell Lethality.
Cancer Cell, 35, 2019
|
|
7SN4
 
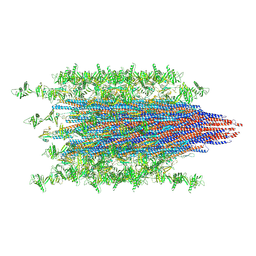 | |
7SN9
 
 | |
8Q4H
 
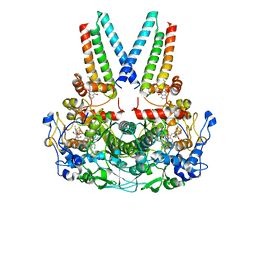 | | a membrane-bound menaquinol:organohalide oxidoreductase complex RDH complex | | Descriptor: | (~{Z})-1,2-bis(chloranyl)ethene, CO-METHYLCOBALAMIN, IRON/SULFUR CLUSTER, ... | | Authors: | Dongchun, N, Ekundayo, B, Henning, S, Julien, M, Holliger, C, Cimmino, L. | | Deposit date: | 2023-08-07 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structure of a membrane-bound menaquinol:organohalide oxidoreductase.
Nat Commun, 14, 2023
|
|
1ZKW
 
 | | Crystal structure of Arg347Ala mutant of botulinum neurotoxin E catalytic domain | | Descriptor: | CHLORIDE ION, ZINC ION, botulinum neurotoxin type E | | Authors: | Agarwal, R, Binz, T, Swaminathan, S. | | Deposit date: | 2005-05-04 | | Release date: | 2005-06-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Analysis of Active Site Residues of Botulinum Neurotoxin E by Mutational, Functional, and Structural Studies: Glu335Gln Is an Apoenzyme.
Biochemistry, 44, 2005
|
|
8POK
 
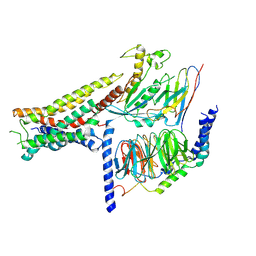 | | Cryo-EM structure of cell-free synthesized human histamine H2 receptor coupled to heterotrimeric Gs protein in lipid environment | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, HISTAMINE, ... | | Authors: | Schnelle, K, Koeck, Z, Persechino, M, Umbach, S, Schihada, H, Januliene, D, Parey, K, Pockes, S, Kolb, P, Doetsch, V, Moeller, A, Hilger, D, Bernhard, F. | | Deposit date: | 2023-07-05 | | Release date: | 2024-03-06 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of cell-free synthesized human histamine 2 receptor/G s complex in nanodisc environment.
Nat Commun, 15, 2024
|
|
4O24
 
 | | DNA Double-Strand Break Repair Pathway Choice Is Directed by Distinct MRE11 Nuclease Activities | | Descriptor: | (5~{Z})-5-[(4-hydroxyphenyl)methylidene]-3-(2-methylpropyl)-2-sulfanylidene-1,3-thiazolidin-4-one, Exonuclease, putative, ... | | Authors: | Shibata, A, Moiani, D, Arvai, A.S, Perry, J, Harding, S.M, Genois, M, Maity, R, Rossum-Fikkert, S, Kertokalio, A, Romoli, F, Ismail, A, Ismalaj, E, Petricci, E, Neale, M.J, Bristow, R.G, Masson, J, Wyman, C, Jeggo, P.A, Tainer, J.A. | | Deposit date: | 2013-12-16 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA Double-Strand Break Repair Pathway Choice Is Directed by Distinct MRE11 Nuclease Activities.
Mol.Cell, 53, 2014
|
|
7ZI0
 
 | | Structure of human Smoothened in complex with cholesterol and SAG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-chloro-N-[trans-4-(methylamino)cyclohexyl]-N-{[3-(pyridin-4-yl)phenyl]methyl}-1-benzothiophene-2-carboxamide, CHOLESTEROL, ... | | Authors: | Byrne, E.F.X, Woolley, R.E, Ansell, B, Sansom, M.S.P, Newstead, S, Siebold, C. | | Deposit date: | 2022-04-07 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Patched 1 regulates Smoothened by controlling sterol binding to its extracellular cysteine-rich domain.
Sci Adv, 8, 2022
|
|
