5DOU
 
 | | Crystal Structure of Human Carbamoyl phosphate synthetase I (CPS1), ligand-bound form | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | de Cima, S, Polo, L.M, Fita, I, Rubio, V. | | Deposit date: | 2015-09-11 | | Release date: | 2015-12-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of human carbamoyl phosphate synthetase: deciphering the on/off switch of human ureagenesis.
Sci Rep, 5, 2015
|
|
5KKN
 
 | | Crystal structure of human ACC2 BC domain in complex with ND-646, the primary amide of ND-630 | | Descriptor: | 2-[1-[(2~{R})-2-(2-methoxyphenyl)-2-(oxan-4-yloxy)ethyl]-5-methyl-6-(1,3-oxazol-2-yl)-2,4-bis(oxidanylidene)thieno[2,3-d]pyrimidin-3-yl]-2-methyl-propanamide, Acetyl-CoA carboxylase 2 | | Authors: | Wang, R, Paul, D, Tong, L. | | Deposit date: | 2016-06-22 | | Release date: | 2016-07-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Acetyl-CoA carboxylase inhibition by ND-630 reduces hepatic steatosis, improves insulin sensitivity, and modulates dyslipidemia in rats.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3VA7
 
 | | Crystal structure of the Kluyveromyces lactis Urea Carboxylase | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, GLYCEROL, KLLA0E08119p, ... | | Authors: | Fan, C, Xiang, S. | | Deposit date: | 2011-12-29 | | Release date: | 2012-02-01 | | Last modified: | 2013-07-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of urea carboxylase provides insights into the carboxyltransfer reaction
J.Biol.Chem., 287, 2012
|
|
3JRW
 
 | | Phosphorylated BC domain of ACC2 | | Descriptor: | Acetyl-CoA carboxylase 2 | | Authors: | Cho, Y.S, Lee, J.I, Shin, D, Kim, H.T, Lee, T.G, Heo, Y.S. | | Deposit date: | 2009-09-09 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular mechanism for the regulation of human ACC2 through phosphorylation by AMPK
Biochem.Biophys.Res.Commun., 391, 2010
|
|
2VR1
 
 | |
6W2J
 
 | | CPS1 bound to allosteric inhibitor H3B-374 | | Descriptor: | (2-fluoranyl-4-methoxy-phenyl)-[(3~{R},5~{R})-4-(2-fluoranyl-4-methoxy-phenyl)carbonyl-3,5-dimethyl-piperazin-1-yl]methanone, 1,2-ETHANEDIOL, Carbamoyl-phosphate synthase [ammonia], ... | | Authors: | Larsen, N.A, Nguyen, T.V. | | Deposit date: | 2020-03-05 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Discovery of 2,6-Dimethylpiperazines as Allosteric Inhibitors of CPS1.
Acs Med.Chem.Lett., 11, 2020
|
|
8GK8
 
 | | R21A Staphylococcus aureus pyruvate carboxylase | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ACETYL COENZYME *A, COENZYME A, ... | | Authors: | Laseke, A.J, St.Maurice, M. | | Deposit date: | 2023-03-17 | | Release date: | 2023-08-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Allosteric Site at the Biotin Carboxylase Dimer Interface Mediates Activation and Inhibition in Staphylococcus aureus Pyruvate Carboxylase.
Biochemistry, 62, 2023
|
|
8FXI
 
 | | Cryo-EM structure of Stanieria sp. CphA2 in complex with ADPCP and 4x(beta-Asp-Arg) | | Descriptor: | 1-[(4-aminopyrimidin-5-yl)amino]-2,5-anhydro-1-deoxy-6-O-[(S)-hydroxy{[(R)-hydroxy(phosphonomethyl)phosphoryl]oxy}phosphoryl]-D-allitol, 4x(beta-Asp-Arg), MAGNESIUM ION, ... | | Authors: | Markus, L.M, Sharon, I, Strauss, M, Schmeing, T.M. | | Deposit date: | 2023-01-24 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure and function of a hexameric cyanophycin synthetase 2.
Protein Sci., 32, 2023
|
|
8J7D
 
 | | Human 3-methylcrotonyl-CoA carboxylase in BCCP-H1 state | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, Methylcrotonoyl-CoA carboxylase beta chain, mitochondrial, ... | | Authors: | Liu, D.S, Su, J.Y. | | Deposit date: | 2023-04-27 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Human 3-methylcrotonyl-CoA carboxylase in BCCP-H1 state
To Be Published
|
|
4HQ6
 
 | | BC domain in the presence of citrate | | Descriptor: | Acetyl-CoA carboxylase 2 | | Authors: | Heo, Y.S. | | Deposit date: | 2012-10-25 | | Release date: | 2013-10-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Insights into the Regulation of ACC2 by Citrate
Bull.Korean Chem.Soc., 34, 2013
|
|
3HBL
 
 | | Crystal Structure of S. aureus Pyruvate Carboxylase T908A Mutant | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ADENOSINE-5'-DIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Tong, L, Yu, L.P.C. | | Deposit date: | 2009-05-04 | | Release date: | 2009-06-30 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | A Symmetrical Tetramer for S. aureus Pyruvate Carboxylase in Complex with Coenzyme A.
Structure, 17, 2009
|
|
8J4Z
 
 | |
8FXH
 
 | |
2GPS
 
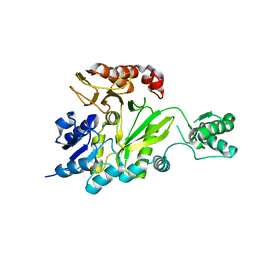 | | Crystal Structure of the Biotin Carboxylase Subunit, E23R mutant, of Acetyl-CoA Carboxylase from Escherichia coli. | | Descriptor: | Biotin carboxylase | | Authors: | Shen, Y, Chou, C.Y, Chang, G.G, Tong, L. | | Deposit date: | 2006-04-18 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Is dimerization required for the catalytic activity of bacterial biotin carboxylase?
Mol.Cell, 22, 2006
|
|
4HNV
 
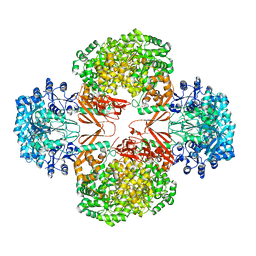 | | Crystal structure of R54E mutant of S. aureus Pyruvate carboxylase | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Yu, L.P.C, Tong, L. | | Deposit date: | 2012-10-21 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Characterizing the Importance of the Biotin Carboxylase Domain Dimer for Staphylococcus aureus Pyruvate Carboxylase Catalysis.
Biochemistry, 52, 2013
|
|
4HNT
 
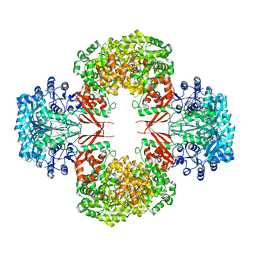 | | crystal structure of F403A mutant of S. aureus Pyruvate carboxylase | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Yu, L.P.C, Tong, L. | | Deposit date: | 2012-10-21 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Characterizing the Importance of the Biotin Carboxylase Domain Dimer for Staphylococcus aureus Pyruvate Carboxylase Catalysis.
Biochemistry, 52, 2013
|
|
3BG5
 
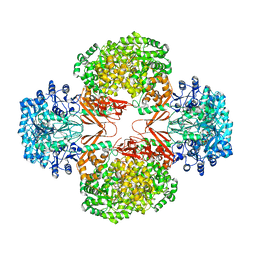 | | Crystal Structure of Staphylococcus Aureus Pyruvate Carboxylase | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Xiang, S, Tong, L. | | Deposit date: | 2007-11-26 | | Release date: | 2008-02-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of human and Staphylococcus aureus pyruvate carboxylase and molecular insights into the carboxyltransfer reaction.
Nat.Struct.Mol.Biol., 15, 2008
|
|
8J99
 
 | | Human 3-methylcrotonyl-CoA carboxylase in BCS-mcoa state | | Descriptor: | Methylcrotonoyl-CoA carboxylase beta chain, mitochondrial, Methylcrotonoyl-CoA carboxylase subunit alpha, ... | | Authors: | Liu, D.S, Su, J.Y. | | Deposit date: | 2023-05-02 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Human 3-methylcrotonyl-CoA carboxylase in BCS-mcoa state
To Be Published
|
|
3HB9
 
 | | Crystal Structure of S. aureus Pyruvate Carboxylase A610T Mutant | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ADENOSINE-5'-DIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Tong, L, Yu, L.P.C. | | Deposit date: | 2009-05-04 | | Release date: | 2009-06-30 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A Symmetrical Tetramer for S. aureus Pyruvate Carboxylase in Complex with Coenzyme A.
Structure, 17, 2009
|
|
3HO8
 
 | |
3U9T
 
 | |
4HNU
 
 | | crystal structure of K442E mutant of S. aureus Pyruvate carboxylase | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ADENOSINE-5'-DIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Yu, L.P.C, Tong, L. | | Deposit date: | 2012-10-21 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Characterizing the Importance of the Biotin Carboxylase Domain Dimer for Staphylococcus aureus Pyruvate Carboxylase Catalysis.
Biochemistry, 52, 2013
|
|
8HZ5
 
 | |
7TA5
 
 | |
5KS8
 
 | |
