8FTD
 
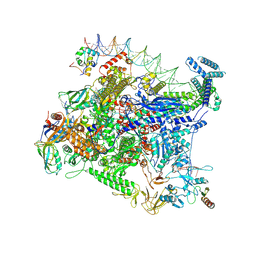 | | Structure of Escherichia coli CedA in complex with transcription initiation complex | | Descriptor: | CHAPSO, Cell division activator CedA, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Liu, M, Vassyliev, N, Nudler, E. | | Deposit date: | 2023-01-11 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | General transcription factor from Escherichia coli with a distinct mechanism of action.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8W8P
 
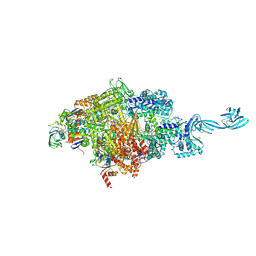 | |
8W8O
 
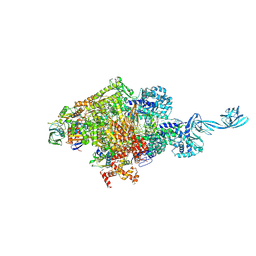 | |
6EEC
 
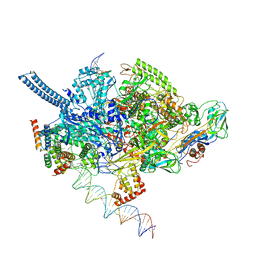 | | Mycobacterium tuberculosis RNAP promoter unwinding intermediate complex with RbpA/CarD and AP3 promoter captured by Corallopyronin | | Descriptor: | DNA (63-MER), DNA (65-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Darst, S.A, Campbell, E.A, Boyaci Selcuk, H, Chen, J. | | Deposit date: | 2018-08-13 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Structures of an RNA polymerase promoter melting intermediate elucidate DNA unwinding.
Nature, 565, 2019
|
|
6EDT
 
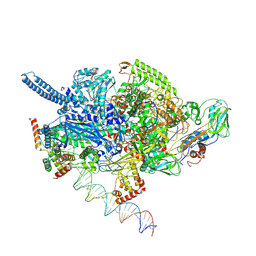 | | Mycobacterium tuberculosis RNAP open promoter complex with RbpA/CarD and AP3 promoter | | Descriptor: | DNA (65-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Darst, S.A, Campbell, E.A, Boyaci Selcuk, H, Chen, J. | | Deposit date: | 2018-08-10 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY | | Cite: | Structures of an RNA polymerase promoter melting intermediate elucidate DNA unwinding.
Nature, 565, 2019
|
|
6EYD
 
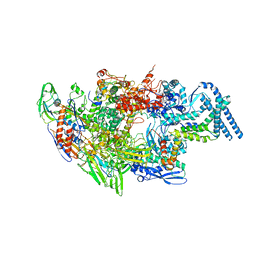 | | Structure of Mycobacterium smegmatis RNA polymerase Sigma-A holoenzyme | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Kouba, T, Barvik, I, Krasny, L. | | Deposit date: | 2017-11-11 | | Release date: | 2018-12-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.22 Å) | | Cite: | The Core and Holoenzyme Forms of RNA Polymerase fromMycobacterium smegmatis.
J. Bacteriol., 201, 2019
|
|
6FBV
 
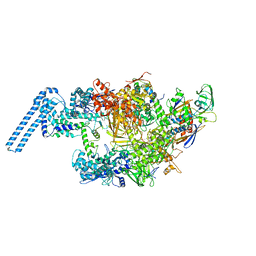 | | Single particle cryo em structure of Mycobacterium tuberculosis RNA polymerase in complex with Fidaxomicin | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Das, K, Lin, W, Ebright, E. | | Deposit date: | 2017-12-19 | | Release date: | 2018-02-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Structural Basis of Transcription Inhibition by Fidaxomicin (Lipiarmycin A3).
Mol. Cell, 70, 2018
|
|
8GZH
 
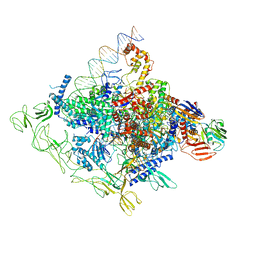 | | Cryo-EM structure of Synechocystis sp. PCC 6803 CTP-bound RPitc | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | You, L.L, Shen, L.Q, Zhang, Y. | | Deposit date: | 2022-09-27 | | Release date: | 2023-04-19 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | An SI3-sigma arch stabilizes cyanobacteria transcription initiation complex.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8GZG
 
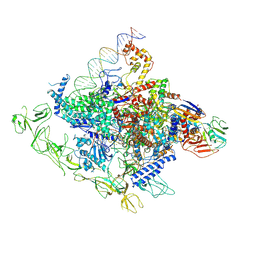 | | Cryo-EM structure of Synechocystis sp. PCC 6803 RPitc | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Shen, L.Q, You, L.L, Zhang, Y. | | Deposit date: | 2022-09-27 | | Release date: | 2023-04-19 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | An SI3-sigma arch stabilizes cyanobacteria transcription initiation complex.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6EE8
 
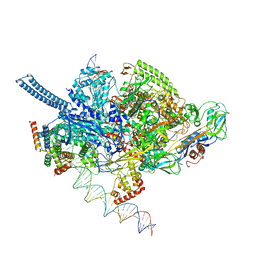 | | Mycobacterium tuberculosis RNAP promoter unwinding intermediate complex with RbpA/CarD and AP3 promoter | | Descriptor: | DNA (60-MER), DNA (65-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Darst, S.A, Campbell, E.A, Boyaci Selcuk, H, Chen, J. | | Deposit date: | 2018-08-13 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Structures of an RNA polymerase promoter melting intermediate elucidate DNA unwinding.
Nature, 565, 2019
|
|
4G7O
 
 | |
4G7Z
 
 | |
7UBM
 
 | |
7UBN
 
 | |
8TOE
 
 | |
8TO1
 
 | | Escherichia coli RNA polymerase unwinding intermediate (I1a) at the lambda PR promoter | | Descriptor: | (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthrene-3,7,12-triol, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Darst, S.A, Saecker, R.M, Mueller, A.U. | | Deposit date: | 2023-08-02 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Early intermediates in bacterial RNA polymerase promoter melting visualized by time-resolved cryo-electron microscopy.
Nat.Struct.Mol.Biol., 2024
|
|
8TO6
 
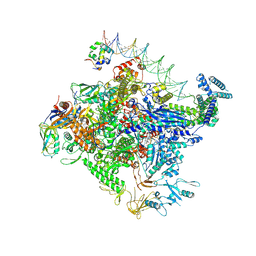 | | Escherichia coli RNA polymerase unwinding intermediate (I1d) at the lambda PR promoter | | Descriptor: | (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthrene-3,7,12-triol, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Darst, S.A, Saecker, R.M, Mueller, A.U. | | Deposit date: | 2023-08-02 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Early intermediates in bacterial RNA polymerase promoter melting visualized by time-resolved cryo-electron microscopy.
Nat.Struct.Mol.Biol., 2024
|
|
8TOM
 
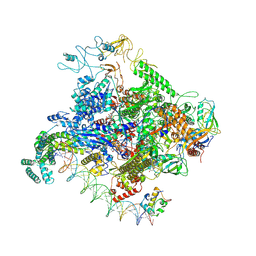 | |
8TO8
 
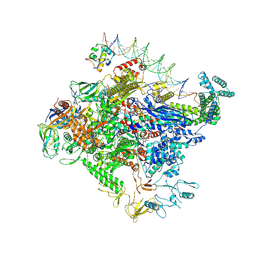 | | Escherichia coli RNA polymerase unwinding intermediate (I1b) at the lambda PR promoter | | Descriptor: | (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthrene-3,7,12-triol, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Darst, S.A, Saecker, R.M, Mueller, A.U. | | Deposit date: | 2023-08-03 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Early intermediates in bacterial RNA polymerase promoter melting visualized by time-resolved cryo-electron microscopy.
Nat.Struct.Mol.Biol., 2024
|
|
8U3B
 
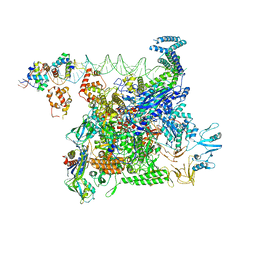 | | Cryo-EM structure of E. coli NarL-transcription activation complex at 3.2A | | Descriptor: | DNA (69-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Liu, B, Kompaniiets, D, Wang, D. | | Deposit date: | 2023-09-07 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural basis for transcription activation by the nitrate-responsive regulator NarL.
Nucleic Acids Res., 52, 2024
|
|
7U22
 
 | | Mycobacterium tuberculosis RNA polymerase sigma A holoenzyme open promoter complex containing UMN-7 | | Descriptor: | (9S,14S,15R,16S,17R,18R,19R,20S,21S,25R)-5,6,18,20-tetrahydroxy-14-methoxy-7,9,15,17,19,21,25-heptamethyl-1'-(2-methylpropyl)-10,26-dioxo-1,3,9,10-tetrahydrospiro[9,4-(epoxypentadecanoimino)furo[2',3':7,8]naphtho[1,2-d]imidazole-2,4'-piperidin]-16-yl benzoate, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Molodtsov, V, Ebright, R.H. | | Deposit date: | 2022-02-22 | | Release date: | 2022-10-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.87 Å) | | Cite: | Redesign of Rifamycin Antibiotics to Overcome ADP-Ribosylation-Mediated Resistance.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
1ZYR
 
 | | Structure of Thermus thermophilus RNA polymerase holoenzyme in complex with the antibiotic streptolydigin | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase omega chain, ... | | Authors: | Tuske, S, Sarafianos, S.G, Wang, X, Hudson, B, Sineva, E, Mukhopadhyay, J, Birktoft, J.J, Leroy, O, Ismail, S, Clark, A.D, Dharia, C, Napoli, A, Laptenko, O, Lee, J, Borukhov, S, Ebright, R.H, Arnold, E. | | Deposit date: | 2005-06-10 | | Release date: | 2005-09-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Inhibition of bacterial RNA polymerase by streptolydigin: stabilization of a straight-bridge-helix active-center conformation.
Cell(Cambridge,Mass.), 122, 2005
|
|
2A6E
 
 | | Crystal structure of the T. Thermophilus RNA polymerase holoenzyme | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Artsimovitch, I, Vassylyeva, M.N, Svetlov, D, Svetlov, V, Perederina, A, Igarashi, N, Matsugaki, N, Wakatsuki, S, Tahirov, T.H, Vassylyev, D.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-02 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Allosteric modulation of the RNA polymerase catalytic reaction is an essential component of transcription control by rifamycins.
Cell(Cambridge,Mass.), 122, 2005
|
|
7VF9
 
 | | Cryo-EM structure of Pseudomonas aeruginosa RNAP sigmaS holoenzyme complexes | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | He, D.W, You, L.L, Zhang, Y. | | Deposit date: | 2021-09-10 | | Release date: | 2022-07-27 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.04 Å) | | Cite: | Pseudomonas aeruginosa SutA wedges RNAP lobe domain open to facilitate promoter DNA unwinding.
Nat Commun, 13, 2022
|
|
7VPD
 
 | |
