4NU8
 
 | |
4O5C
 
 | |
3BX2
 
 | | Puf4 RNA binding domain bound to HO endonuclease RNA 3' UTR recognition sequence | | Descriptor: | HO endonuclease 3' UTR binding sequence, Protein PUF4, SODIUM ION, ... | | Authors: | Miller, M.T, Higgin, J.J, Hall, T.M.T. | | Deposit date: | 2008-01-11 | | Release date: | 2008-03-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Basis of altered RNA-binding specificity by PUF proteins revealed by crystal structures of yeast Puf4p
Nat.Struct.Mol.Biol., 15, 2008
|
|
4F63
 
 | | Crystal structure of Human Fibroblast Growth Factor Receptor 1 Kinase domain in complex with compound 1 | | Descriptor: | 1,2-ETHANEDIOL, 5-bromo-N~4~-(3-methyl-1H-pyrazol-5-yl)-N~2~-[2-(pyridin-3-yl)ethyl]pyrimidine-2,4-diamine, Fibroblast growth factor receptor 1 | | Authors: | Norman, R.A, Breed, J, Ogg, D. | | Deposit date: | 2012-05-14 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Protein-Ligand Crystal Structures Can Guide the Design of Selective Inhibitors of the FGFR Tyrosine Kinase.
J.Med.Chem., 55, 2012
|
|
4F64
 
 | | Crystal structure of Human Fibroblast Growth Factor Receptor 1 Kinase domain in complex with compound 6 | | Descriptor: | 1,2-ETHANEDIOL, 5-bromo-N~4~-[3-(3-methoxypropyl)-1H-pyrazol-5-yl]-N~2~-[(3-methyl-1,2-oxazol-5-yl)methyl]pyrimidine-2,4-diamine, Fibroblast growth factor receptor 1, ... | | Authors: | Norman, R.A, Breed, J, Ogg, D. | | Deposit date: | 2012-05-14 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Protein-Ligand Crystal Structures Can Guide the Design of Selective Inhibitors of the FGFR Tyrosine Kinase.
J.Med.Chem., 55, 2012
|
|
4NXB
 
 | | Crystal structure of iLOV-I486(2LT) at pH 7.0 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | Authors: | Wang, J, Li, J, Liu, X. | | Deposit date: | 2013-12-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.561 Å) | | Cite: | Significant expansion of fluorescent protein sensing ability through the genetic incorporation of superior photo-induced electron-transfer quenchers.
J.Am.Chem.Soc., 136, 2014
|
|
4NXL
 
 | | Dibenzothiophene monooxygenase (DszC) from Rhodococcus erythropolis | | Descriptor: | DszC | | Authors: | Zhang, L, Duan, X, Li, X, Rao, Z. | | Deposit date: | 2013-12-09 | | Release date: | 2014-07-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the stabilization of active, tetrameric DszC by its C-terminus.
Proteins, 82, 2014
|
|
4FAZ
 
 | |
4O21
 
 | | Product complex of metal-free PKAc, ATP-gamma-S and SP20. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Thio-phosphorylated peptide pSP20, cAMP-dependent protein kinase catalytic subunit alpha | | Authors: | Das, A, Kovalevsky, A.Y, Gerlits, O, Langan, P, Heller, W.T, Keshwani, M, Taylor, S. | | Deposit date: | 2013-12-16 | | Release date: | 2014-05-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Metal-Free cAMP-Dependent Protein Kinase Can Catalyze Phosphoryl Transfer.
Biochemistry, 53, 2014
|
|
3BX3
 
 | |
3BZK
 
 | |
3GV6
 
 | | Crystal Structure of human chromobox homolog 6 (CBX6) with H3K9 peptide | | Descriptor: | Chromobox protein homolog 6, Histone H3K9me3 peptide | | Authors: | Dong, A, Amaya, M.F, Li, Z, Loppnau, P, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-03-30 | | Release date: | 2009-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Recognition and specificity determinants of the human cbx chromodomains.
J.Biol.Chem., 286, 2011
|
|
4O72
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with NU7441 | | Descriptor: | 1,2-ETHANEDIOL, 8-(dibenzo[b,d]thiophen-4-yl)-2-(morpholin-4-yl)-4H-chromen-4-one, Bromodomain-containing protein 4, ... | | Authors: | Zhu, J.-Y, Ember, S.W, Watts, C, Schonbrunn, E. | | Deposit date: | 2013-12-24 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Acetyl-lysine Binding Site of Bromodomain-Containing Protein 4 (BRD4) Interacts with Diverse Kinase Inhibitors.
Acs Chem.Biol., 9, 2014
|
|
4FFJ
 
 | | The crystal structure of spDHBPs from S.pneumoniae | | Descriptor: | GLYCEROL, Riboflavin biosynthesis protein ribBA, SULFATE ION | | Authors: | Wang, D. | | Deposit date: | 2012-06-01 | | Release date: | 2013-07-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The crystal structure of SpDHBPs from S. pneumoniae
To be Published
|
|
4F0K
 
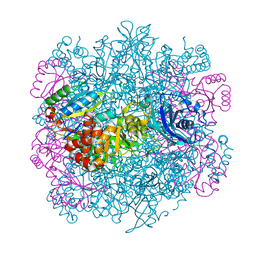 | | UNACTIVATED RUBISCO with MAGNESIUM AND CARBON DIOXIDE BOUND | | Descriptor: | CARBON DIOXIDE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stec, B. | | Deposit date: | 2012-05-04 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural mechanism of RuBisCO activation by carbamylation of the active site lysine.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4OI6
 
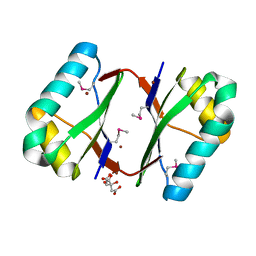 | | Crystal structure analysis of nickel-bound form SCO4226 from Streptomyces coelicolor A3(2) | | Descriptor: | CITRIC ACID, NICKEL (II) ION, Nickel responsive protein | | Authors: | Lu, M, Jiang, Y.L, Wang, S, Cheng, W, Zhang, R.G, Virolle, M.J, Chen, Y, Zhou, C.Z. | | Deposit date: | 2014-01-18 | | Release date: | 2014-09-10 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Streptomyces coelicolor SCO4226 Is a Nickel Binding Protein.
Plos One, 9, 2014
|
|
4FQO
 
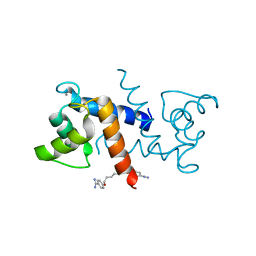 | | Crystal Structure of Calcium-Loaded S100B Bound to SBi4211 | | Descriptor: | 4,4'-[heptane-1,7-diylbis(oxy)]dibenzenecarboximidamide, CALCIUM ION, Protein S100-B | | Authors: | McKnight, L.E, Raman, E.P, Bezawada, P, Kudrimoti, S, Wilder, P.T, Hartman, K.G, Toth, E.A, Coop, A, MacKerrell, A.D, Weber, D.J. | | Deposit date: | 2012-06-25 | | Release date: | 2012-10-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-Based Discovery of a Novel Pentamidine-Related Inhibitor of the Calcium-Binding Protein S100B.
ACS Med Chem Lett, 3, 2012
|
|
4V0Z
 
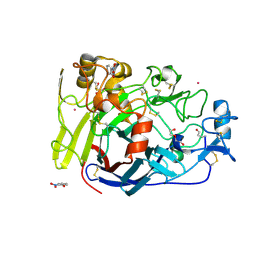 | |
4V1W
 
 | |
7Y74
 
 | |
7YA3
 
 | |
6X1H
 
 | |
6X2D
 
 | | Crystal Structure of DNase I Domain of Ribonuclease E from Vibrio cholerae | | Descriptor: | IODIDE ION, Ribonuclease E | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Wiersum, G, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-05-20 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of DNase I Domain of Ribonuclease E from Vibrio cholerae.
To Be Published
|
|
4V5V
 
 | | Structure of respiratory syncytial virus nucleocapsid protein, P1 crystal form | | Descriptor: | RESPIRATORY SYNCYTIAL VIRUS NUCLEOCAPSID PROTEIN, RNA | | Authors: | El Omari, K, Dhaliwal, B, Ren, J, Abrescia, N.G.A, Lockyer, M, Powell, K.L, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2011-05-04 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structures of Respiratory Syncytial Virus Nucleocapsid Protein from Two Crystal Forms: Details of Potential Packing Interactions in the Native Helical Form.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
6WDW
 
 | |
