6A5E
 
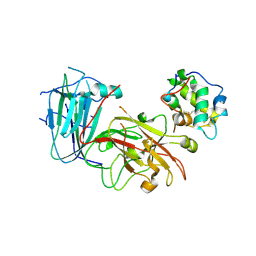 | | Crystal structure of plant peptide RALF23 in complex with FER and LLG2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GPI-anchored protein LLG2, ... | | Authors: | Xiao, Y, Chai, J. | | Deposit date: | 2018-06-23 | | Release date: | 2019-06-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.766 Å) | | Cite: | Mechanisms of RALF peptide perception by a heterotypic receptor complex.
Nature, 572, 2019
|
|
6A8I
 
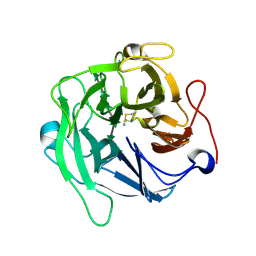 | | Crystal structure of endo-arabinanase ABN-TS D147N mutant in complex with arabinohexaose | | Descriptor: | CALCIUM ION, alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose, endo-alpha-(1->5)-L-arabinanase | | Authors: | Yamaguchi, A, Tada, T. | | Deposit date: | 2018-07-09 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of endo-1,5-alpha-L-arabinanase mutants from Bacillus thermodenitrificans TS-3 in complex with arabino-oligosaccharides.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
5ZSC
 
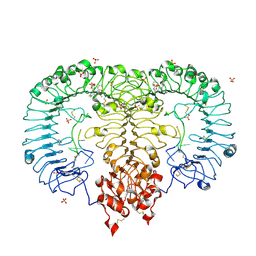 | | Crystal structure of monkey TLR7 in complex with IMDQ and CCUUCC | | Descriptor: | 1-[[4-(aminomethyl)phenyl]methyl]-2-butyl-imidazo[4,5-c]quinolin-4-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2018-04-28 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Analyses of Toll-like Receptor 7 Reveal Detailed RNA Sequence Specificity and Recognition Mechanism of Agonistic Ligands.
Cell Rep, 25, 2018
|
|
8HMS
 
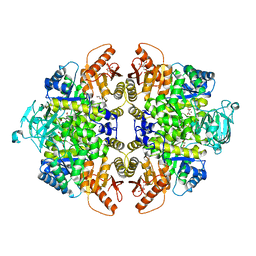 | | Crystal Structure of PKM2 mutant C474S | | Descriptor: | 1,2-ETHANEDIOL, 1,6-di-O-phosphono-beta-D-fructofuranose, GLYCEROL, ... | | Authors: | Upadhyay, S, Kumar, A, Patel, A.K. | | Deposit date: | 2022-12-05 | | Release date: | 2023-01-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and mechanistic insights into cancer patient-derived mutations in Pyruvate Kinase muscle isoform 2
To Be Published
|
|
2BNM
 
 | |
2BSH
 
 | |
6JE7
 
 | | Crystal structure of human serum albumin crystallized by ammonium sulfate | | Descriptor: | Serum albumin | | Authors: | Kitahara, M, Fudo, S, Yoneda, T, Nukaga, M, Hoshino, T. | | Deposit date: | 2019-02-04 | | Release date: | 2019-03-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Anisotropic Distribution of Ammonium Sulfate Ions in Protein Crystallization
Cryst.Growth Des., 19, 2019
|
|
2BZG
 
 | | Crystal structure of thiopurine S-methyltransferase. | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, S-ADENOSYL-L-HOMOCYSTEINE, THIOPURINE S-METHYLTRANSFERASE | | Authors: | Battaile, K.P, Wu, H, Zeng, H, Loppnau, P, Dong, A, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-08-17 | | Release date: | 2005-08-25 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural Basis of Allele Variation of Human Thiopurine-S-Methyltransferase.
Proteins, 67, 2007
|
|
8HMQ
 
 | | Crystal Structure of PKM2 mutant P403A | | Descriptor: | 1,2-ETHANEDIOL, 1,6-di-O-phosphono-beta-D-fructofuranose, GLYCEROL, ... | | Authors: | Upadhyay, S, Kumar, A, Patel, A.K. | | Deposit date: | 2022-12-05 | | Release date: | 2023-01-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and mechanistic insights into cancer patient-derived mutations in Pyruvate Kinase muscle isoform 2
To Be Published
|
|
2C2C
 
 | |
2RTI
 
 | | STREPTAVIDIN-GLYCOLURIL COMPLEX, PH 2.50, SPACE GROUP I222 | | Descriptor: | FORMIC ACID, GLYCOLURIL, STREPTAVIDIN | | Authors: | Katz, B.A. | | Deposit date: | 1997-09-11 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Binding of biotin to streptavidin stabilizes intersubunit salt bridges between Asp61 and His87 at low pH.
J.Mol.Biol., 274, 1997
|
|
2RJI
 
 | |
8GZC
 
 | | Crystal structure of EP300 HAT domain in complex with compound 10 | | Descriptor: | (2~{R},4~{R})-4-fluoranyl-1-[1-(4-methoxyphenyl)cyclohexyl]carbonyl-~{N}-(1~{H}-pyrazolo[4,3-b]pyridin-5-yl)pyrrolidine-2-carboxamide, Histone acetyltransferase p300, ZINC ION | | Authors: | Takahashi, M, Hanzawa, H. | | Deposit date: | 2022-09-26 | | Release date: | 2023-01-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of DS-9300: A Highly Potent, Selective, and Once-Daily Oral EP300/CBP Histone Acetyltransferase Inhibitor.
J.Med.Chem., 66, 2023
|
|
8HMR
 
 | | Crystal Structure of PKM2 mutant L144P | | Descriptor: | 1,2-ETHANEDIOL, 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, ... | | Authors: | Upadhyay, S, Kumar, A, Patel, A.K. | | Deposit date: | 2022-12-05 | | Release date: | 2023-01-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and mechanistic insights into cancer patient-derived mutations in Pyruvate Kinase muscle isoform 2
To Be Published
|
|
2RFC
 
 | | Ligand bound (4-phenylimidazole) Crystal Structure of a Cytochrome P450 from the Thermoacidophilic Archaeon Picrophilus Torridus | | Descriptor: | 4-PHENYL-1H-IMIDAZOLE, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ho, W.W, Li, H, Poulos, T.L, Nishida, C.R, Ortiz de Montellano, P.R. | | Deposit date: | 2007-09-28 | | Release date: | 2008-01-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure and Properties of CYP231A2 from the Thermoacidophilic Archaeon Picrophilus torridus.
Biochemistry, 47, 2008
|
|
8HC5
 
 | | SARS-CoV-2 wildtype S1 in complex with YB9-258 Fab and R1-32 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32 Fab, Heavy chain of YB9-258 Fab, ... | | Authors: | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-25 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
8RWZ
 
 | | Open non-crosslinked structure Brd4BD2-MZ1-(NEDD8)-CRL2VHL | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-2-[2-[2-[2-[2-[2-[(9~{S})-7-(4-chlorophenyl)-4,5,13-trimethyl-3-thia-1,8,11,12-tetrazatricyclo[8.3.0.0^{2,6}]trideca-2(6),4,7,10,12-pentaen-9-yl]ethanoylamino]ethoxy]ethoxy]ethoxy]ethanoylamino]-3,3-dimethyl-butanoyl]-~{N}-[[4-(4-methyl-2,3-dihydro-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Bromodomain-containing protein 4, Cullin-2, ... | | Authors: | Crowe, C, Nakasone, M.A, Ciulli, A. | | Deposit date: | 2024-02-05 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Open non-crosslinked structure Brd4BD2-MZ1-(NEDD8)-CRL2VHL
To Be Published
|
|
2BSR
 
 | | Crystal structures and KIR3DL1 recognition of three immunodominant viral peptides complexed to HLA-B2705 | | Descriptor: | BETA-2-MICROGLOBULIN, EPSTEIN-BARR NUCLEAR ANTIGEN-6, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Stewart-Jones, G.B.E, Di Gleria, K, Kollnberger, S, Mcmichael, A.J, Jones, E.Y, Bowness, P. | | Deposit date: | 2005-05-23 | | Release date: | 2005-05-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures and Kir3Dl1 Recognition of Three Immunodominant Viral Peptides Complexed to Hla-B2705
Eur.J.Immunol., 35, 2005
|
|
8H1A
 
 | |
2RG9
 
 | | Crystal structure of viscum album mistletoe lectin I in native state at 1.95 A resolution, comparison of structure active site conformation in ricin and in viscumin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AZIDE ION, Beta-galactoside-specific lectin 1 chain A isoform 1, ... | | Authors: | Karpechenko, N.U, Timofeev, V.I, Gabdoulkhakov, A.G, Mikhailov, A.M. | | Deposit date: | 2007-10-03 | | Release date: | 2008-10-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of viscum album mistletoe lectin I in native state at 1.95 A resolution, comparison of structure active site conformation in ricin and in viscumin
To be Published
|
|
2RLI
 
 | | Solution structure of Cu(I) human Sco2 | | Descriptor: | COPPER (I) ION, SCO2 protein homolog, mitochondrial | | Authors: | Banci, L, Bertini, I, Ciofi-baffoni, S, Gerothanassis, I.P, Leontari, I, Martinelli, M, Wang, S, Structural Proteomics in Europe (SPINE), Structural Proteomics in Europe 2 (SPINE-2) | | Deposit date: | 2007-07-11 | | Release date: | 2007-08-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A Structural Characterization of Human SCO2
Structure, 15, 2007
|
|
6JM5
 
 | | Crystal structure of TBC1D23 C terminal domain | | Descriptor: | SODIUM ION, TBC1 domain family member 23 | | Authors: | Sun, Q, Huang, W. | | Deposit date: | 2019-03-07 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional studies of TBC1D23 C-terminal domain provide a link between endosomal trafficking and PCH.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6JR4
 
 | | Crystal structure of a NIR-emitting DNA-stabilized Ag16 nanocluster | | Descriptor: | CALCIUM ION, DNA (5'-D(*CP*AP*CP*CP*TP*AP*GP*CP*GP*A)-3'), SILVER ION | | Authors: | Kondo, J, Cerretani, C, Kanazawa, H, Vosch, T. | | Deposit date: | 2019-04-02 | | Release date: | 2019-08-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Crystal structure of a NIR-Emitting DNA-Stabilized Ag16Nanocluster.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
8HMU
 
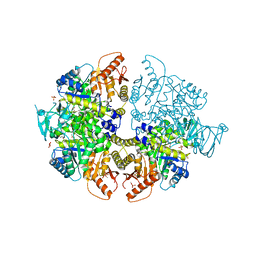 | | Crystal Structure of PKM2 mutant R516C | | Descriptor: | 1,2-ETHANEDIOL, 1,6-di-O-phosphono-beta-D-fructofuranose, GLYCEROL, ... | | Authors: | Upadhyay, S, Kumar, A, Patel, A.K. | | Deposit date: | 2022-12-05 | | Release date: | 2023-01-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and mechanistic insights into cancer patient-derived mutations in Pyruvate Kinase muscle isoform 2
To Be Published
|
|
2RHI
 
 | |
