1UM9
 
 | | branched-chain 2-oxo acid dehydrogenase (E1) from Thermus thermophilus HB8 in apo-form | | Descriptor: | 2-oxo acid dehydrogenase alpha subunit, 2-oxo acid dehydrogenase beta subunit, SULFATE ION | | Authors: | Nakai, T, Nakagawa, N, Maoka, N, Masui, R, Kuramitsu, S, Kamiya, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-25 | | Release date: | 2004-03-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ligand-induced Conformational Changes and a Reaction Intermediate in Branched-chain 2-Oxo Acid Dehydrogenase (E1) from Thermus thermophilus HB8, as Revealed by X-ray Crystallography
J.Mol.Biol., 337, 2004
|
|
4MNW
 
 | | Crystal structure of urokinase-type plasminogen activator (uPA) complexed with bicyclic peptide UK749 | | Descriptor: | 1,3,5-tris(bromomethyl)benzene, ACETATE ION, GLYCEROL, ... | | Authors: | Chen, S, Pojer, F, Heinis, C. | | Deposit date: | 2013-09-11 | | Release date: | 2014-02-05 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Peptide ligands stabilized by small molecules.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
5OO0
 
 | | Cdk2(WT) covalent adduct with D28 at C177 | | Descriptor: | Cyclin-dependent kinase 2, methyl 4-propanoyl-2,3-dihydroquinoxaline-1-carboxylate | | Authors: | Craven, G, Morgan, R.M.L, Mann, D.J. | | Deposit date: | 2017-08-04 | | Release date: | 2018-03-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-Throughput Kinetic Analysis for Target-Directed Covalent Ligand Discovery.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
4MNY
 
 | | Crystal structure of urokinase-type plasminogen activator (uPA) complexed with bicyclic peptide UK903 | | Descriptor: | ACETATE ION, GLYCEROL, N,N',N''-benzene-1,3,5-triyltris(2-bromoacetamide), ... | | Authors: | Chen, S, Pojer, F, Heinis, C. | | Deposit date: | 2013-09-11 | | Release date: | 2014-02-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Peptide ligands stabilized by small molecules.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
5V6Y
 
 | |
2J0D
 
 | |
5HVM
 
 | | Structure of Aspergillus fumigatus trehalose-6-phosphate synthase A in complex with UDP and validoxylamine A | | Descriptor: | (1S,2S,3R,6S)-4-(HYDROXYMETHYL)-6-{[(1S,2S,3S,4R,5R)-2,3,4-TRIHYDROXY-5-(HYDROXYMETHYL)CYCLOHEXYL]AMINO}CYCLOHEX-4-ENE-1,2,3-TRIOL, Alpha,alpha-trehalose-phosphate synthase (UDP-forming), URIDINE-5'-DIPHOSPHATE | | Authors: | Miao, Y, Brennan, R.G. | | Deposit date: | 2016-01-28 | | Release date: | 2017-05-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.815 Å) | | Cite: | Structural and In Vivo Studies on Trehalose-6-Phosphate Synthase from Pathogenic Fungi Provide Insights into Its Catalytic Mechanism, Biological Necessity, and Potential for Novel Antifungal Drug Design.
MBio, 8, 2017
|
|
3PCJ
 
 | | STRUCTURE OF PROTOCATECHUATE 3,4-DIOXYGENASE COMPLEXED WITH 2-HYDROXYISONICOTINIC ACID N-OXIDE | | Descriptor: | 2-HYDROXYISONICOTINIC ACID N-OXIDE, BETA-MERCAPTOETHANOL, FE (III) ION, ... | | Authors: | Orville, A.M, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 1997-07-18 | | Release date: | 1998-01-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structures of substrate and substrate analog complexes of protocatechuate 3,4-dioxygenase: endogenous Fe3+ ligand displacement in response to substrate binding.
Biochemistry, 36, 1997
|
|
3PCL
 
 | | STRUCTURE OF PROTOCATECHUATE 3,4-DIOXYGENASE COMPLEXED WITH 2-HYDROXYISONICOTINIC ACID N-OXIDE AND CYANIDE | | Descriptor: | 2-HYDROXYISONICOTINIC ACID N-OXIDE, CYANIDE ION, FE (III) ION, ... | | Authors: | Orville, A.M, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 1997-07-18 | | Release date: | 1998-01-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures of substrate and substrate analog complexes of protocatechuate 3,4-dioxygenase: endogenous Fe3+ ligand displacement in response to substrate binding.
Biochemistry, 36, 1997
|
|
3PCA
 
 | | STRUCTURE OF PROTOCATECHUATE 3,4-DIOXYGENASE COMPLEXED WITH 3,4-DIHYDROXYBENZOATE | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, BETA-MERCAPTOETHANOL, FE (III) ION, ... | | Authors: | Orville, A.M, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 1997-07-18 | | Release date: | 1998-01-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of substrate and substrate analog complexes of protocatechuate 3,4-dioxygenase: endogenous Fe3+ ligand displacement in response to substrate binding.
Biochemistry, 36, 1997
|
|
3PCM
 
 | | STRUCTURE OF PROTOCATECHUATE 3,4-DIOXYGENASE COMPLEXED WITH 6-HYDROXYNICOTINIC ACID N-OXIDE AND CYANIDE | | Descriptor: | 6-HYDROXYISONICOTINIC ACID N-OXIDE, CYANIDE ION, FE (III) ION, ... | | Authors: | Orville, A.M, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 1997-07-18 | | Release date: | 1998-01-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structures of substrate and substrate analog complexes of protocatechuate 3,4-dioxygenase: endogenous Fe3+ ligand displacement in response to substrate binding.
Biochemistry, 36, 1997
|
|
3PCK
 
 | | STRUCTURE OF PROTOCATECHUATE 3,4-DIOXYGENASE COMPLEXED WITH 6-HYDROXYNICOTINIC ACID N-OXIDE | | Descriptor: | 6-HYDROXYISONICOTINIC ACID N-OXIDE, BETA-MERCAPTOETHANOL, FE (III) ION, ... | | Authors: | Orville, A.M, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 1997-07-18 | | Release date: | 1998-01-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structures of substrate and substrate analog complexes of protocatechuate 3,4-dioxygenase: endogenous Fe3+ ligand displacement in response to substrate binding.
Biochemistry, 36, 1997
|
|
5HRV
 
 | | Crystal structure of the fifth bromodomain of human PB1 in complex with 1-ethylisochromeno[3,4-c]pyrazol-5(2H)-one) compound | | Descriptor: | 1,2-ETHANEDIOL, 1-ethylisochromeno[3,4-c]pyrazol-5(3H)-one, Protein polybromo-1 | | Authors: | Tallant, C, Myrianthopoulos, V, Gaboriaud-Kolar, N, Newman, J.A, Picaud, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Mikros, E, Knapp, S. | | Deposit date: | 2016-01-24 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery and Optimization of a Selective Ligand for the Switch/Sucrose Nonfermenting-Related Bromodomains of Polybromo Protein-1 by the Use of Virtual Screening and Hydration Analysis.
J.Med.Chem., 59, 2016
|
|
7OVH
 
 | | Crystal structure of the VIM-2 acquired metallo-beta-Lactamase in Complex with compound 14 (JMV-6931) | | Descriptor: | ACETATE ION, Metallo-beta-lactamase VIM-2-like protein, UNKNOWN LIGAND, ... | | Authors: | Tassone, G, Benvenuti, M, Verdirosa, F, Sannio, F, Marcoccia, F, Docquier, J.D, Pozzi, C, Mangani, S. | | Deposit date: | 2021-06-14 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1,2,4-Triazole-3-thione compounds with a 4-ethyl alkyl/aryl sulfide substituent are broad-spectrum metallo-beta-lactamase inhibitors with re-sensitization activity.
Eur.J.Med.Chem., 226, 2021
|
|
7OVE
 
 | | Crystal structure of the VIM-2 acquired metallo-beta-Lactamase in Complex with compound 10 (JMV-7210) | | Descriptor: | ACETATE ION, Metallo-beta-lactamase VIM-2-like protein, UNKNOWN LIGAND, ... | | Authors: | Tassone, G, Benvenuti, M, Verdirosa, F, Sannio, F, Mangani, S, Docquier, J.D, Pozzi, C, Marcoccia, F. | | Deposit date: | 2021-06-14 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | 1,2,4-Triazole-3-thione compounds with a 4-ethyl alkyl/aryl sulfide substituent are broad-spectrum metallo-beta-lactamase inhibitors with re-sensitization activity.
Eur.J.Med.Chem., 226, 2021
|
|
8EBM
 
 | |
8EBL
 
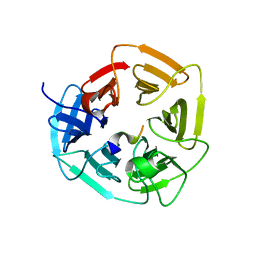 | |
8QNG
 
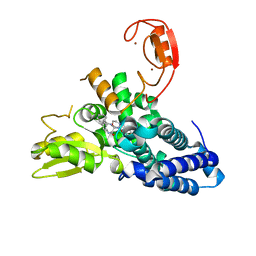 | |
8QNI
 
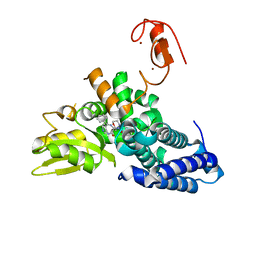 | |
8QNH
 
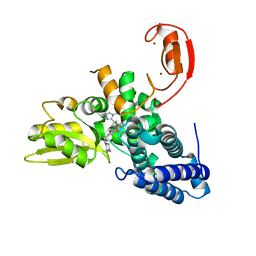 | | Crystal structure of the E3 ubiquitin ligase Cbl-b with an allosteric inhibitor (WO2020264398 Ex23) | | Descriptor: | 2-{3-[(1s,3R)-3-methyl-1-(4-methyl-4H-1,2,4-triazol-3-yl)cyclobutyl]phenyl}-6-{[(3S)-3-methylpiperidin-1-yl]methyl}-4-(trifluoromethyl)-2,3-dihydro-1H-isoindol-1-one, E3 ubiquitin-protein ligase CBL-B, SODIUM ION, ... | | Authors: | Schimpl, M. | | Deposit date: | 2023-09-26 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Discovery of a Novel Benzodiazepine Series of Cbl-b Inhibitors for the Enhancement of Antitumor Immunity.
Acs Med.Chem.Lett., 14, 2023
|
|
4PUN
 
 | | tRNA-Guanine Transglycosylase (TGT) Apo-Structure pH 7.8 | | Descriptor: | DIMETHYL SULFOXIDE, Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Neeb, M, Heine, A, Klebe, G. | | Deposit date: | 2014-03-13 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Chasing Protons: How Isothermal Titration Calorimetry, Mutagenesis, and pKa Calculations Trace the Locus of Charge in Ligand Binding to a tRNA-Binding Enzyme.
J.Med.Chem., 57, 2014
|
|
6CK4
 
 | | G96A mutant of the PRPP riboswitch from T. mathranii bound to ppGpp | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GUANOSINE-5',3'-TETRAPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Reiss, C.W, Knappenberger, A.J, Strobel, S.A. | | Deposit date: | 2018-02-27 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.097 Å) | | Cite: | Structures of two aptamers with differing ligand specificity reveal ruggedness in the functional landscape of RNA.
Elife, 7, 2018
|
|
5OVQ
 
 | |
4H6O
 
 | | Sterol 14-alpha demethylase (CYP51)from Trypanosoma cruzi in complex with the inhibitor NEU321 (1-(3-(4-chloro-3,5-dimethylphenoxy)benzyl)-1H-imidazole | | Descriptor: | 1-[3-(4-chloro-3,5-dimethylphenoxy)benzyl]-1H-imidazole, PROTOPORPHYRIN IX CONTAINING FE, Sterol 14-alpha demethylase | | Authors: | Wawrzak, Z, Hargrove, T.Y, Pollastri, M.P, Lepesheva, G.I. | | Deposit date: | 2012-09-19 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Antitrypanosomal Lead Discovery: Identification of a Ligand-Efficient Inhibitor of Trypanosoma cruzi CYP51 and Parasite Growth.
J.Med.Chem., 56, 2013
|
|
1N5Z
 
 | | Complex structure of Pex13p SH3 domain with a peptide of Pex14p | | Descriptor: | 14-mer peptide from Peroxisomal membrane protein PEX14, Peroxisomal membrane protein PAS20 | | Authors: | Douangamath, A, Filipp, F.V, Klein, A.T.J, Barnett, P, Zou, P, Voorn-Brouwer, T, Vega, M.C, Mayans, O.M, Sattler, M, Distel, B, Wilmanns, M. | | Deposit date: | 2002-11-08 | | Release date: | 2002-12-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Topography for Independent Binding of alpha-Helical and PPII-Helical Ligands to a Peroxisomal SH3 Domain
MOL.CELL, 10, 2002
|
|
