6MVL
 
 | |
5BJU
 
 | | X-ray structure of the PglF dehydratase from Campylobacter jejuni in complex with UDP and NAD(H) | | Descriptor: | 1,2-ETHANEDIOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Riegert, A.S, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-09-12 | | Release date: | 2017-11-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Biochemical Investigation of PglF from Campylobacter jejuni Reveals a New Mechanism for a Member of the Short Chain Dehydrogenase/Reductase Superfamily.
Biochemistry, 56, 2017
|
|
1X26
 
 | | Solution structure of the AA-mismatch DNA complexed with naphthyridine-azaquinolone | | Descriptor: | 5'-D(*CP*AP*TP*TP*CP*AP*GP*TP*TP*AP*G)-3', 5'-D(*CP*TP*AP*AP*CP*AP*GP*AP*AP*TP*G)-3', N~3~-{3-[(7-METHYL-1,8-NAPHTHYRIDIN-2-YL)AMINO]-3-OXOPROPYL}-N~1~-[(7-OXO-7,8-DIHYDRO-1,8-NAPHTHYRIDIN-2-YL)METHYL]-BET A-ALANINAMIDE | | Authors: | Nakatani, K, Hagihara, S, Goto, Y, Kobori, A, Hagihara, M, Hayashi, G, Kyo, M, Nomura, M, Mishima, M, Kojima, C. | | Deposit date: | 2005-04-20 | | Release date: | 2006-04-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Small-molecule ligand induces nucleotide flipping in (CAG)n trinucleotide repeats
Nat.Chem.Biol., 1, 2005
|
|
2Y43
 
 | |
6JH7
 
 | | Crystal structure of AerF from Microcystis aeruginosa | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, Short chain dehydrogenase family protein, ... | | Authors: | Qiu, X. | | Deposit date: | 2019-02-17 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structural and functional investigation of AerF, a NADPH-dependent alkenal double bond reductase participating in the biosynthesis of Choi moiety of aeruginosin
J.Struct.Biol., 2019
|
|
1HEX
 
 | |
4XT1
 
 | | Structure of a nanobody-bound viral GPCR bound to human chemokine CX3CL1 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHOLESTEROL, Fractalkine, ... | | Authors: | Burg, J.S, Jude, K.M, Waghray, D, Garcia, K.C. | | Deposit date: | 2015-01-22 | | Release date: | 2015-03-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.886 Å) | | Cite: | Structural biology. Structural basis for chemokine recognition and activation of a viral G protein-coupled receptor.
Science, 347, 2015
|
|
6JHB
 
 | |
1WGM
 
 | | Solution structure of the U-box in human ubiquitin conjugation factor E4A | | Descriptor: | Ubiquitin conjugation factor E4A | | Authors: | Saito, K, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the U-box in human ubiquitin conjugation factor E4A
To be Published
|
|
6JHA
 
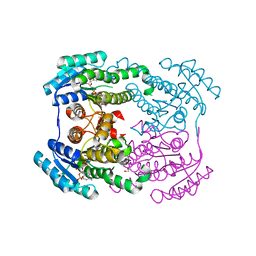 | | Crystal structure of NADPH bound AerF from Microcystis aeruginosa | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short chain dehydrogenase family protein, TRIETHYLENE GLYCOL | | Authors: | Qiu, X. | | Deposit date: | 2019-02-17 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural and functional investigation of AerF, a NADPH-dependent alkenal double bond reductase participating in the biosynthesis of Choi moiety of aeruginosin
J.Struct.Biol., 2019
|
|
2Y93
 
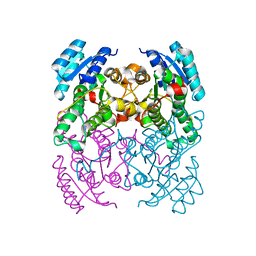 | | Crystal Structure of cis-Biphenyl-2,3-dihydrodiol-2,3-dehydrogenase (BphB)from Pandoraea pnomenusa strain B-356. | | Descriptor: | CIS-2,3-DIHYDROBIPHENYL-2,3-DIOL DEHYDROGENASE | | Authors: | Dhindwal, S, Patil, D.N, Kumar, P. | | Deposit date: | 2011-02-11 | | Release date: | 2011-08-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Biochemical Studies and Ligand-Bound Structures of Biphenyl Dehydrogenase from Pandoraea Pnomenusa Strain B-356 Reveal a Basis for Broad Specificity of the Enzyme.
J.Biol.Chem., 286, 2011
|
|
1WZ0
 
 | | Solution Structure of Human SUMO-2 (SMT3B), a Ubiquitin-like Protein | | Descriptor: | Ubiquitin-like protein SMT3B | | Authors: | Zhao, C, Saito, K, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-02-21 | | Release date: | 2005-08-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Human SUMO-2 (SMT3B), a Ubiquitin-like Protein
To be Published
|
|
4ZSC
 
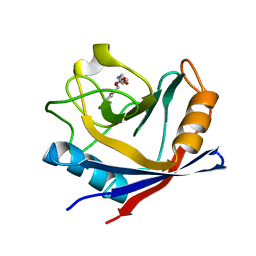 | | Human Cyclophilin D Complexed with an Inhibitor at room temperature | | Descriptor: | Peptidyl-prolyl cis-trans isomerase F, mitochondrial, ethyl N-[(4-aminobenzyl)carbamoyl]glycinate | | Authors: | Gelin, M, Delfosse, V, Allemand, F, Hoh, F, Sallaz-Damaz, Y, Pirocchi, M, Bourguet, W, Ferrer, J.-L, Labesse, G, Guichou, J.-F. | | Deposit date: | 2015-05-13 | | Release date: | 2015-08-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Combining `dry' co-crystallization and in situ diffraction to facilitate ligand screening by X-ray crystallography.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7T9X
 
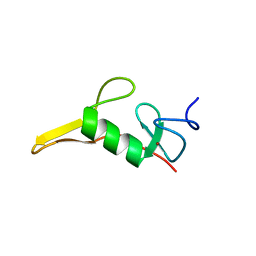 | | Saccharomyces cerevisiae Pex12 RING domain | | Descriptor: | Peroxisome assembly protein 12, ZINC ION | | Authors: | Feng, P, Rapoport, T. | | Deposit date: | 2021-12-20 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | A peroxisomal ubiquitin ligase complex forms a retrotranslocation channel.
Nature, 607, 2022
|
|
2Y99
 
 | | Crystal Structure of cis-Biphenyl-2,3-dihydrodiol-2,3-dehydrogenase (BphB)from Pandoraea pnomenusa strain B-356 complex with co-enzyme NAD | | Descriptor: | CIS-2,3-DIHYDROBIPHENYL-2,3-DIOL DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Dhindwal, S, Patil, D.N, Kumar, P. | | Deposit date: | 2011-02-12 | | Release date: | 2011-08-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Biochemical Studies and Ligand-Bound Structures of Biphenyl Dehydrogenase from Pandoraea Pnomenusa Strain B-356 Reveal a Basis for Broad Specificity of the Enzyme.
J.Biol.Chem., 286, 2011
|
|
6Y98
 
 | | Crystal Structure of subtype-switched Epithelial Adhesin 9 to 1 A domain (Epa9-CBL2Epa1) from Candida glabrata in complex with beta-lactose | | Descriptor: | CALCIUM ION, PA14 domain-containing protein, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Hoffmann, D, Diderrich, R, Kock, M, Friederichs, S, Reithofer, V, Essen, L.-O, Moesch, H.-U. | | Deposit date: | 2020-03-06 | | Release date: | 2020-07-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Functional reprogramming ofCandida glabrataepithelial adhesins: the role of conserved and variable structural motifs in ligand binding.
J.Biol.Chem., 295, 2020
|
|
3QLW
 
 | | Candida albicans dihydrofolate reductase complexed with NADPH and 5-[3-(2,5-dimethoxyphenyl)prop-1-yn-1-yl]-6-ethylpyrimidine-2,4-diamine (UCP120B) | | Descriptor: | 5-[3-(2,5-dimethoxyphenyl)prop-1-yn-1-yl]-6-ethylpyrimidine-2,4-diamine, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative uncharacterized protein CaJ7.0360 | | Authors: | Paulsen, J.L, Bendel, S.D, Anderson, A.C. | | Deposit date: | 2011-02-03 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | Crystal Structures of Candida albicans Dihydrofolate Reductase Bound to Propargyl-Linked Antifolates Reveal the Flexibility of Active Site Loop Residues Critical for Ligand Potency and Selectivity.
Chem.Biol.Drug Des., 78, 2011
|
|
6Y9J
 
 | | Crystal Structure of subtype-switched Epithelial Adhesin 1 to 9 A domain (Epa1-CBL2Epa9) from Candida glabrata in complex with beta-lactose | | Descriptor: | CALCIUM ION, CHLORIDE ION, Epa1p, ... | | Authors: | Hoffmann, D, Diderrich, R, Kock, M, Friederichs, S, Reithofer, V, Essen, L.-O, Moesch, H.-U. | | Deposit date: | 2020-03-09 | | Release date: | 2020-07-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Functional reprogramming ofCandida glabrataepithelial adhesins: the role of conserved and variable structural motifs in ligand binding.
J.Biol.Chem., 295, 2020
|
|
1XB7
 
 | | X-ray structure of ERRalpha LBD in complex with a PGC-1alpha peptide at 2.5A resolution | | Descriptor: | IODIDE ION, Peroxisome proliferator activated receptor gamma coactivator 1 alpha, Steroid hormone receptor ERR1 | | Authors: | Kallen, J, Schlaeppi, J.M, Bitsch, F, Filipuzzi, I, Schilb, A, Riou, V, Graham, A, Strauss, A, Geiser, M, Fournier, B. | | Deposit date: | 2004-08-30 | | Release date: | 2004-09-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Evidence for Ligand-independent Transcriptional Activation of the Human Estrogen-related Receptor {alpha} (ERR{alpha}): CRYSTAL STRUCTURE OF ERR{alpha} LIGAND BINDING DOMAIN IN COMPLEX WITH PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR COACTIVATOR-1{alpha}
J.Biol.Chem., 279, 2004
|
|
6NBP
 
 | | Crystal Structure of a Sugar N-Formyltransferase from the Plant Pathogen Pantoea ananatis | | Descriptor: | N-formyltransferase, N-{[4-({[(6R)-2-amino-5-formyl-4-oxo-1,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)phenyl]carbonyl}-L-glutamic acid, PHOSPHATE ION, ... | | Authors: | Hofmeister, D.L, Thoden, J.B, Holden, H.M. | | Deposit date: | 2018-12-09 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Investigation of a sugar N-formyltransferase from the plant pathogen Pantoea ananatis.
Protein Sci., 28, 2019
|
|
8QBN
 
 | |
8QE8
 
 | | Structure of the non-canonical CTLH E3 substrate receptor WDR26 bound to NMNAT1 substrate | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, Nicotinamide/nicotinic acid mononucleotide adenylyltransferase 1, WD repeat-containing protein 26, ... | | Authors: | Chrustowicz, J, Sherpa, D, Schulman, B.A. | | Deposit date: | 2023-08-30 | | Release date: | 2024-05-15 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Non-canonical substrate recognition by the human WDR26-CTLH E3 ligase regulates prodrug metabolism.
Mol.Cell, 84, 2024
|
|
6CR2
 
 | | Crystal structure of sterol 14-alpha demethylase (CYP51B) from Aspergillus fumigatus in complex with the VNI derivative N-(1-(2,4-dichlorophenyl)-2-(1H-imidazol-1-yl)ethyl)-4-(5-(2-fluoro-4-(2,2,2-trifluoroethoxy)phenyl)-1,3,4-oxadiazol-2-yl)benzamide | | Descriptor: | 14-alpha sterol demethylase Cyp51B, N-[(1R)-1-(2,4-dichlorophenyl)-2-(1H-imidazol-1-yl)ethyl]-4-{5-[2-fluoro-4-(2,2,2-trifluoroethoxy)phenyl]-1,3,4-oxadiazol-2-yl}benzamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Friggeri, L, Hargrove, T.Y, Wawrzak, Z, Lepesheva, G.I. | | Deposit date: | 2018-03-16 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Sterol 14 alpha-Demethylase Structure-Based Design of VNI (( R)- N-(1-(2,4-Dichlorophenyl)-2-(1 H-imidazol-1-yl)ethyl)-4-(5-phenyl-1,3,4-oxadiazol-2-yl)benzamide)) Derivatives To Target Fungal Infections: Synthesis, Biological Evaluation, and Crystallographic Analysis.
J. Med. Chem., 61, 2018
|
|
1W9S
 
 | |
3UIW
 
 | | Zebrafish Grx2 (APO) | | Descriptor: | GLUTATHIONE, Glutaredoxin 2, SULFATE ION | | Authors: | McDonough, M.A, Johansson, C. | | Deposit date: | 2011-11-06 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | A New Mode of Iron-sulfur Cluster Coordination in Glutaredoxins is Crucial for Axonogenesis
To be Published
|
|
