4LV4
 
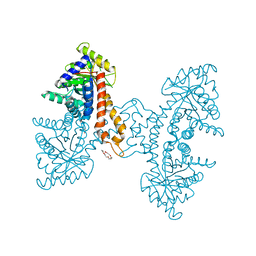 | | A noncompetitive inhibitor for M. tuberculosis's class IIa fructose 1,6-bisphosphate aldolase | | Descriptor: | 8-hydroxyquinoline-2-carboxylic acid, ACETATE ION, Fructose-bisphosphate aldolase, ... | | Authors: | Capodagli, G.C, Pegan, S.D. | | Deposit date: | 2013-07-25 | | Release date: | 2014-01-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | A Noncompetitive Inhibitor for Mycobacterium tuberculosis's Class IIa Fructose 1,6-Bisphosphate Aldolase.
Biochemistry, 53, 2014
|
|
4LU4
 
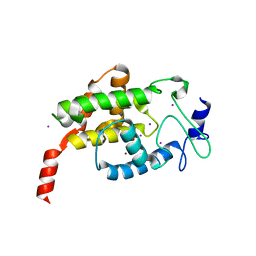 | |
4LYF
 
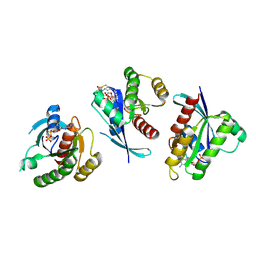 | | Crystal Structure of small molecule vinylsulfonamide 8 covalently bound to K-Ras G12C | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, N-{1-[N-(4,5-dichloro-2-hydroxyphenyl)glycyl]piperidin-4-yl}ethanesulfonamide | | Authors: | Ostrem, J.M, Peters, U, Sos, M.L, Wells, J.A, Shokat, K.M. | | Deposit date: | 2013-07-31 | | Release date: | 2013-11-27 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.568 Å) | | Cite: | K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions.
Nature, 503, 2013
|
|
4LYW
 
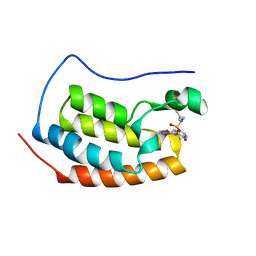 | | Crystal Structure of BRD4(1) bound to inhibitor XD14 | | Descriptor: | 4-acetyl-N-[5-(diethylsulfamoyl)-2-hydroxyphenyl]-3-ethyl-5-methyl-1H-pyrrole-2-carboxamide, Bromodomain-containing protein 4 | | Authors: | Wohlwend, D, Gerhardt, S, Einsle, O. | | Deposit date: | 2013-07-31 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 4-Acyl pyrroles: mimicking acetylated lysines in histone code reading.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
4DLR
 
 | | H-Ras PEG 400/Ca(OAc)2, ordered off | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, CALCIUM ION, GTPase HRas, ... | | Authors: | Holzapfel, G, Mattos, C. | | Deposit date: | 2012-02-06 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.319 Å) | | Cite: | Shift in the Equilibrium between On and Off States of the Allosteric Switch in Ras-GppNHp Affected by Small Molecules and Bulk Solvent Composition.
Biochemistry, 51, 2012
|
|
4DLY
 
 | | Set 1 CaCl2/DTT, ordered off | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CALCIUM ION, GTPase HRas, ... | | Authors: | Holzapfel, G, Mattos, C. | | Deposit date: | 2012-02-06 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Shift in the Equilibrium between On and Off States of the Allosteric Switch in Ras-GppNHp Affected by Small Molecules and Bulk Solvent Composition.
Biochemistry, 51, 2012
|
|
2QDM
 
 | |
2QEB
 
 | | Crystal Structure of Anopheles Gambiae D7R4-Histamine Complex | | Descriptor: | D7R4 Protein, GLYCEROL, HISTAMINE | | Authors: | Andersen, J.F, Mans, B.J, Calvo, E, Ribeiro, J.M. | | Deposit date: | 2007-06-25 | | Release date: | 2007-10-09 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | The Crystal Structure of D7r4, a Salivary Biogenic Amine-binding Protein from the Malaria Mosquito Anopheles gambiae
J.Biol.Chem., 282, 2007
|
|
4DI2
 
 | | Crystal structure of BACE1 in complex with hydroxyethylamine inhibitor 37 | | Descriptor: | (2R)-N-{(2S,3R)-4-{[(4'S)-6'-(2,2-dimethylpropyl)-3',4'-dihydrospiro[cyclobutane-1,2'-pyrano[2,3-b]pyridin]-4'-yl]amino}-3-hydroxy-1-[3-(1,3-thiazol-2-yl)phenyl]butan-2-yl}-2-methoxypropanamide, Beta-secretase 1, GLYCEROL | | Authors: | Whittington, D.A, Long, A.M. | | Deposit date: | 2012-01-30 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and synthesis of potent, orally efficacious hydroxyethylamine derived beta-site amyloid precursor protein cleaving enzyme (BACE1) inhibitors.
J.Med.Chem., 55, 2012
|
|
4DPH
 
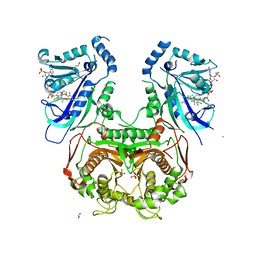 | | Quadruple mutant (N51I+C59R+S108N+I164L) Plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS) complexed with P65 and NADPH | | Descriptor: | 2,4-diamino-6-methyl-5-[3-(2,4,5-trichlorophenoxy)propyloxy]pyrimidine, BETA-MERCAPTOETHANOL, Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | Authors: | Yuthavong, Y, Vilaivan, T, Kamchonwongpaisan, S, Charman, S.A, McLennan, D.N, White, K.L, Vivas, L, Bongard, E, Chitnumsub, P, Tarnchompoo, B, Thongphanchang, C, Taweechai, S, Vanichtanakul, J, Arwon, U, Fantauzzi, P, Yuvaniyama, J, Charman, W.N, Matthews, D. | | Deposit date: | 2012-02-13 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Malarial dihydrofolate reductase as a paradigm for drug development against a resistance-compromised target
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4DQ5
 
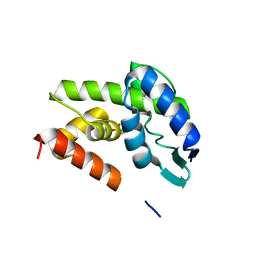 | | Structural Investigation of Bacteriophage Phi6 Lysin (WT) | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, Membrane protein Phi6 P5wt | | Authors: | Dessau, M.A, Modis, Y. | | Deposit date: | 2012-02-15 | | Release date: | 2013-01-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.395 Å) | | Cite: | Selective pressure causes an RNA virus to trade reproductive fitness for increased structural and thermal stability of a viral enzyme.
PLoS Genet, 8, 2012
|
|
4DJG
 
 | |
4DQF
 
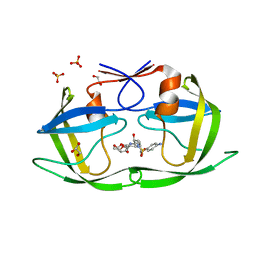 | | Crystal Structure of (G16A/L38A) HIV-1 Protease in Complex with DRV | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, 1,2-ETHANEDIOL, Aspartyl protease, ... | | Authors: | Schiffer, C.A, Mittal, S. | | Deposit date: | 2012-02-15 | | Release date: | 2012-03-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Hydrophobic core flexibility modulates enzyme activity in HIV-1 protease.
J.Am.Chem.Soc., 134, 2012
|
|
2QH7
 
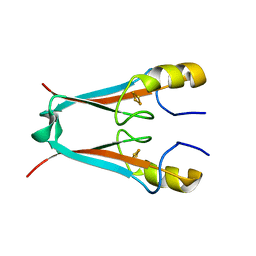 | | MitoNEET is a uniquely folded 2Fe-2S outer mitochondrial membrane protein stabilized by pioglitazone | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Zinc finger CDGSH-type domain 1 | | Authors: | Paddock, M.L, Wiley, S.E, Axelrod, H.L, Cohen, A.E, Roy, M, Abresch, E.C, Capraro, D, Murphy, A.N, Nechushtai, R, Dixon, J.E, Jennings, P.A. | | Deposit date: | 2007-06-30 | | Release date: | 2007-08-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | MitoNEET is a uniquely folded 2Fe 2S outer mitochondrial membrane protein stabilized by pioglitazone.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
4DLF
 
 | | Crystal structure of an amidohydrolase (COG3618) from burkholderia multivorans (TARGET EFI-500235) with bound ZN, space group P3221 | | Descriptor: | Amidohydrolase 2, GLYCEROL, UNKNOWN ATOM OR ION, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Seidel, R.D, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Al Obaidi, N.F, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Raushel, F.M, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-02-06 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.925 Å) | | Cite: | Crystal structure of an amidohydrolase (COG3618) from burkholderia multivorans (target efi-500235) with bound ZN, space group P3221
to be published
|
|
4DR4
 
 | | Crystal structure of the Thermus thermophilus (HB8) 30S ribosomal subunit with codon, cognate transfer RNA anticodon stem-loop and multiple copies of paromomycin molecules bound | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Demirci, H, Murphy IV, F, Murphy, E, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2012-02-16 | | Release date: | 2012-11-14 | | Last modified: | 2013-01-30 | | Method: | X-RAY DIFFRACTION (3.969 Å) | | Cite: | A structural basis for streptomycin-induced misreading of the genetic code.
Nat Commun, 4, 2013
|
|
2QOB
 
 | | Human EphA3 kinase domain, base structure | | Descriptor: | BETA-MERCAPTOETHANOL, Ephrin receptor | | Authors: | Davis, T, Walker, J.R, Newman, E.M, Mackenzie, F, Butler-Cole, C, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-07-20 | | Release date: | 2007-08-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Autoregulation by the Juxtamembrane Region of the Human Ephrin Receptor Tyrosine Kinase A3 (EphA3).
Structure, 16, 2008
|
|
4DTY
 
 | | cytochrome P450 BM3h-8C8 MRI sensor, no ligand | | Descriptor: | MAGNESIUM ION, PROTOPORPHYRIN IX CONTAINING FE, cytochrome P450 BM3 variant 8C8 | | Authors: | Brustad, E.M, Lelyveld, V.S, Snow, C.D, Crook, N, Martinez, F.M, Scholl, T.J, Jasanoff, A, Arnold, F.H. | | Deposit date: | 2012-02-21 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-guided directed evolution of highly selective p450-based magnetic resonance imaging sensors for dopamine and serotonin.
J.Mol.Biol., 422, 2012
|
|
2QOO
 
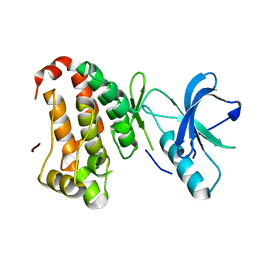 | | Human EphA3 kinase and juxtamembrane region, Y596F:Y602F:Y742F triple mutant | | Descriptor: | Ephrin receptor | | Authors: | Davis, T, Walker, J.R, Newman, E.M, Mackenzie, F, Butler-Cole, C, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-07-20 | | Release date: | 2007-08-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Autoregulation by the Juxtamembrane Region of the Human Ephrin Receptor Tyrosine Kinase A3 (EphA3).
Structure, 16, 2008
|
|
2QIV
 
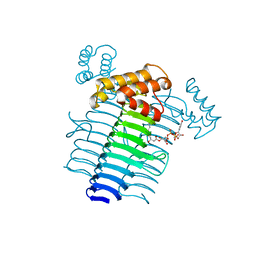 | |
2QJ6
 
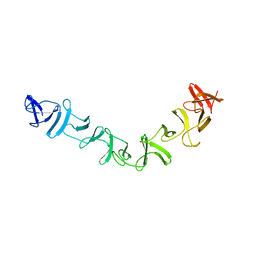 | | Crystal structure analysis of a 14 repeat C-terminal fragment of toxin TcdA in Clostridium difficile | | Descriptor: | Toxin A | | Authors: | Albesa-Jove, D, Bertrand, T, Carpenter, L, Lim, J, Brown, K.A, Fairweather, N. | | Deposit date: | 2007-07-06 | | Release date: | 2008-07-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Solution and crystal structures of the cell binding domain of toxins TcdA and TcdB from Clostridium difficile
To be Published
|
|
4DOI
 
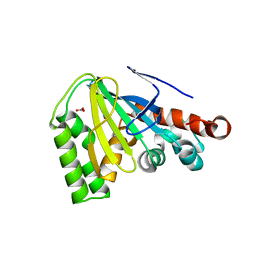 | |
2QP9
 
 | | Crystal Structure of S.cerevisiae Vps4 | | Descriptor: | CADMIUM ION, SULFATE ION, Vacuolar protein sorting-associated protein 4 | | Authors: | Xiao, J, Xu, Z. | | Deposit date: | 2007-07-23 | | Release date: | 2007-10-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural characterization of the ATPase reaction cycle of endosomal AAA protein Vps4.
J.Mol.Biol., 374, 2007
|
|
4DWH
 
 | | Structure of the major type 1 pilus subunit FIMA bound to the FIMC (2.5 A resolution) | | Descriptor: | Chaperone protein fimC, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Scharer, M.A, Puorger, C, Crespo, M, Glockshuber, R, Capitani, G. | | Deposit date: | 2012-02-24 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Quality control of disulfide bond formation in pilus subunits by the chaperone FimC.
Nat.Chem.Biol., 8, 2012
|
|
4DR7
 
 | | Crystal structure of the Thermus thermophilus (HB8) 30S ribosomal subunit with codon, crystallographically disordered near-cognate transfer RNA anticodon stem-loop mismatched at the second codon position, and streptomycin bound | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Demirci, H, Murphy IV, F, Murphy, E, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2012-02-16 | | Release date: | 2012-11-14 | | Last modified: | 2013-01-30 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | A structural basis for streptomycin-induced misreading of the genetic code.
Nat Commun, 4, 2013
|
|
