8B4J
 
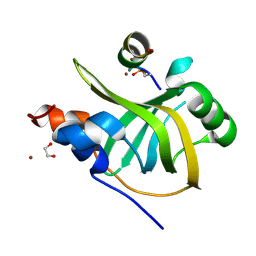 | | Rfa1-N-terminal domain in complex with phosphorylated Ddc2 | | Descriptor: | 1,2-ETHANEDIOL, DNA damage checkpoint protein LCD1, Replication factor A protein 1, ... | | Authors: | Yates, L.A, Zhang, X. | | Deposit date: | 2022-09-20 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | A DNA damage-induced phosphorylation circuit enhances Mec1 ATR Ddc2 ATRIP recruitment to Replication Protein A.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8VCC
 
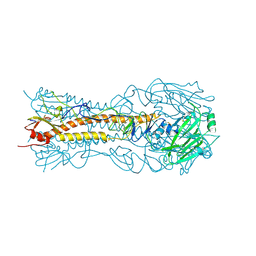 | |
8DK0
 
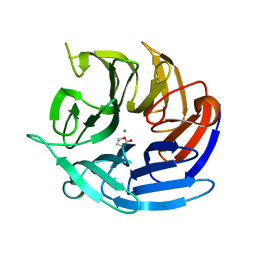 | | Crystal structure of RPA3624, a beta-propeller lactonase from Rhodopseudomonas palustris, with active-site bound (S)gamma-valerolactone | | Descriptor: | CALCIUM ION, Gluconolactonase, SODIUM ION, ... | | Authors: | Bingman, C.A, Hall, B.W, Smith, R.W, Fox, B.G, Donohue, T.J. | | Deposit date: | 2022-07-01 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A broad specificity beta-propeller enzyme from Rhodopseudomonas palustris that hydrolyzes many lactones including gamma-valerolactone.
J.Biol.Chem., 299, 2022
|
|
8DJF
 
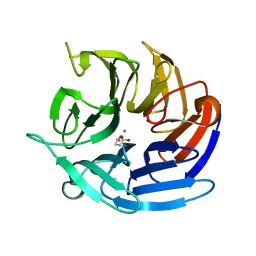 | | Crystal structure of RPA3624, a beta-propeller lactonase from Rhodopseudomonas palustris, with active-site bound tetrahedral intermediate | | Descriptor: | (5S)-5-methyloxolane-2,2-diol, CALCIUM ION, Gluconolactonase, ... | | Authors: | Bingman, C.A, Hall, B.W, Smith, R.W, Fox, B.G, Donohue, T.J. | | Deposit date: | 2022-06-30 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A broad specificity beta-propeller enzyme from Rhodopseudomonas palustris that hydrolyzes many lactones including gamma-valerolactone.
J.Biol.Chem., 299, 2022
|
|
8DJZ
 
 | | Crystal structure of RPA3624, a beta-propeller lactonase from Rhodopseudomonas palustris, with active-site bound product | | Descriptor: | (4R)-4-hydroxypentanoic acid, CALCIUM ION, Gluconolactonase, ... | | Authors: | Bingman, C.A, Hall, B.W, Smith, R.W, Fox, B.G, Donohue, T.J. | | Deposit date: | 2022-07-01 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A broad specificity beta-propeller enzyme from Rhodopseudomonas palustris that hydrolyzes many lactones including gamma-valerolactone.
J.Biol.Chem., 299, 2022
|
|
7A6Q
 
 | | Crystal structure of human aldehyde dehydrogenase 1A3 in complex with selective NR6 inhibitor compound | | Descriptor: | (3-oxidanylidene-3-sodiooxy-propanoyl)oxysodium, 3-(2-phenylimidazo[1,2-a]pyridin-6-yl)benzenecarbonitrile, Aldehyde dehydrogenase family 1 member A3, ... | | Authors: | Gelardi, E.L.M, Garavaglia, S. | | Deposit date: | 2020-08-26 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | A Selective Competitive Inhibitor of Aldehyde Dehydrogenase 1A3 Hinders Cancer Cell Growth, Invasiveness and Stemness In Vitro.
Cancers (Basel), 13, 2021
|
|
6XPQ
 
 | |
6XPX
 
 | |
8V9R
 
 | | Cryo-EM Structure of a Proteolytic ClpXP AAA+ Machine Poised to Unfold a Branched-Degron DHFR-ssrA Substrate Bound with MTX | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, ... | | Authors: | Ghanbarpour, A, Sauer, R.T, Davis, J.H. | | Deposit date: | 2023-12-09 | | Release date: | 2024-10-16 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | A proteolytic AAA+ machine poised to unfold protein substrates.
Nat Commun, 15, 2024
|
|
6XQ4
 
 | |
6XPR
 
 | | Human antibody D2 H1-1/H3-1 H3 in complex with the influenza hemagglutinin head domain of A/Texas/50/2012(H3N2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | McCarthy, K.R, Harrison, S.C, Lee, J. | | Deposit date: | 2020-07-08 | | Release date: | 2021-05-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (4.092 Å) | | Cite: | A Prevalent Focused Human Antibody Response to the Influenza Virus Hemagglutinin Head Interface.
Mbio, 12, 2021
|
|
6G8J
 
 | | 14-3-3sigma in complex with a A130beta3A mutated YAP pS127 phosphopeptide | | Descriptor: | 14-3-3 protein sigma, ACE-ARG-ALA-HIS-SEP-SER-PRO-BAL-SER-LEU-GLN, CHLORIDE ION, ... | | Authors: | Andrei, S.A, Thijssen, V, Brunsveld, L, Ottmann, C, Milroy, L.G. | | Deposit date: | 2018-04-09 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | A study on the effect of synthetic alpha-to-beta3-amino acid mutations on the binding of phosphopeptides to 14-3-3 proteins.
Chem.Commun.(Camb.), 55, 2019
|
|
8VSK
 
 | | Crystal structure of Dehaloperoxidase A in complex with substrate 2,4-dibromophenol | | Descriptor: | 2,4-bis(bromanyl)phenol, DI(HYDROXYETHYL)ETHER, Dehaloperoxidase A, ... | | Authors: | Aktar, M.S, de Serrano, V.S, Ghiladi, R.A, Franzen, S. | | Deposit date: | 2024-01-24 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.515 Å) | | Cite: | Structural Comparison of Substrate Binding Sites in Dehaloperoxidase A and B.
Biochemistry, 63, 2024
|
|
8VZR
 
 | | Crystal structure of dehaloperoxidase A in complex with substrate 4-bromo-o-cresol | | Descriptor: | 4-bromo-2-methylphenol, DI(HYDROXYETHYL)ETHER, Dehaloperoxidase A, ... | | Authors: | Aktar, M.S, de Serrano, V.S, Ghiladi, R.A, Franzen, S. | | Deposit date: | 2024-02-12 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural Comparison of Substrate Binding Sites in Dehaloperoxidase A and B.
Biochemistry, 63, 2024
|
|
6V9T
 
 | | Tudor domain of TDRD3 in complex with a small molecule | | Descriptor: | 4-methyl-2,3,4,5,6,7-hexahydrodicyclopenta[b,e]pyridin-8(1H)-imine, Tudor domain-containing protein 3, UNKNOWN ATOM OR ION | | Authors: | Li, W, Tempel, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-12-16 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.154 Å) | | Cite: | A small molecule antagonist of SMN disrupts the interaction between SMN and RNAP II.
Nat Commun, 13, 2022
|
|
7TRJ
 
 | | The eukaryotic translation initiation factor 2B from Homo sapiens with a H160D mutation in the beta subunit | | Descriptor: | Translation initiation factor eIF-2B subunit alpha, Translation initiation factor eIF-2B subunit beta, Translation initiation factor eIF-2B subunit delta, ... | | Authors: | Wang, L, Schoof, M, Lawrence, R, Boone, M, Frost, A, Walter, P. | | Deposit date: | 2022-01-29 | | Release date: | 2022-04-27 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | A point mutation in the nucleotide exchange factor eIF2B constitutively activates the integrated stress response by allosteric modulation.
Elife, 11, 2022
|
|
6Y1T
 
 | | The crystal structure of engineered cytochrome c peroxidase from Saccharomyces cerevisiae with a Trp51 to S-Trp51 modification | | Descriptor: | 1,2-ETHANEDIOL, Cytochrome c peroxidase, mitochondrial, ... | | Authors: | Ortmayer, M, Levy, C, Green, A.P. | | Deposit date: | 2020-02-13 | | Release date: | 2021-06-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Noncanonical Tryptophan Analogue Reveals an Active Site Hydrogen Bond Controlling Ferryl Reactivity in a Heme Peroxidase.
Jacs Au, 1, 2021
|
|
6Z5J
 
 | | Arrangement of the matrix protein M1 in influenza A/Hong Kong/1/1968 VLPs (HA,NA,M1,M2) | | Descriptor: | Matrix protein 1 | | Authors: | Peukes, J, Xiong, X, Erlendsson, S, Qu, K, Wan, W, Kraeusslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2020-05-26 | | Release date: | 2020-10-14 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | The native structure of the assembled matrix protein 1 of influenza A virus.
Nature, 587, 2020
|
|
4YYH
 
 | | Crystal structure of BRD9 Bromodomain bound to a crotonyllysine peptide | | Descriptor: | Bromodomain-containing protein 9, Histone H4 | | Authors: | Tang, Y, Bellon, S, Cochran, A.G, Poy, F. | | Deposit date: | 2015-03-23 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | A Subset of Human Bromodomains Recognizes Butyryllysine and Crotonyllysine Histone Peptide Modifications.
Structure, 23, 2015
|
|
4YYN
 
 | | Crystal structure of TAF1 BD2 Bromodomain bound to a crotonyllysine peptide | | Descriptor: | Histone H4, Transcription initiation factor TFIID subunit 1 | | Authors: | Tang, Y, Bellon, S, Cochran, A.G, Poy, F. | | Deposit date: | 2015-03-24 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Subset of Human Bromodomains Recognizes Butyryllysine and Crotonyllysine Histone Peptide Modifications.
Structure, 23, 2015
|
|
6XP9
 
 | |
4YYK
 
 | | Crystal structure of BRD9 Bromodomain bound to a crotonyllysine peptide | | Descriptor: | Bromodomain-containing protein 9, Histone H4 | | Authors: | Tang, Y, Bellon, S, Cochran, A.G, Poy, F. | | Deposit date: | 2015-03-24 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A Subset of Human Bromodomains Recognizes Butyryllysine and Crotonyllysine Histone Peptide Modifications.
Structure, 23, 2015
|
|
6Z5L
 
 | |
6Z83
 
 | | CK2 alpha bound to chemical probe SGC-CK2-1 | | Descriptor: | Casein kinase II subunit alpha, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Kraemer, A, Wells, C, Drewry, D.H, Pickett, J.E, Axtman, A.D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-02 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.171 Å) | | Cite: | Development of a potent and selective chemical probe for the pleiotropic kinase CK2.
Cell Chem Biol, 28, 2021
|
|
8VKC
 
 | | Crystal structure of dehaloperoxidase A in complex with substrate 4-nitrophenol | | Descriptor: | Dehaloperoxidase A, GLYCEROL, P-NITROPHENOL, ... | | Authors: | Aktar, M.S, de Serrano, V.S, Ghiladi, R.A, Franzen, S. | | Deposit date: | 2024-01-08 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural Comparison of Substrate Binding Sites in Dehaloperoxidase A and B.
Biochemistry, 63, 2024
|
|
