1XFO
 
 | |
4B2C
 
 | | Structure of the factor Xa-like trypsin variant triple-Ala (TPA) in complex with eglin C | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CATIONIC TRYPSIN, ... | | Authors: | Menzel, A, Neumann, P, Stubbs, M.T. | | Deposit date: | 2012-07-13 | | Release date: | 2012-08-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Thermodynamic signatures in macromolecular interactions involving conformational flexibility.
Biol.Chem., 395, 2014
|
|
3WLC
 
 | | Crystal structure of dimeric GCaMP6m | | Descriptor: | CALCIUM ION, Myosin light chain kinase, Green fluorescent protein, ... | | Authors: | Ding, J, Luo, A.F, Hu, L.Y, Wang, D.C, Shao, F. | | Deposit date: | 2013-11-08 | | Release date: | 2014-01-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural basis of the ultrasensitive calcium indicator GCaMP6.
Sci China Life Sci, 57, 2014
|
|
8U52
 
 | | Co-crystal structure of human transthyretin conjugated to the stilbene substructure derived from reaction with the fluorogenic covalent kinetic stabilizer A2 | | Descriptor: | S-(4-fluorophenyl) 3-(dimethylamino)-5-[(E)-2-(4-hydroxy-3,5-dimethylphenyl)ethenyl]benzenecarbothioate, Transthyretin | | Authors: | Yan, N.L, Nugroho, K, Kline, G.M, Basanta, B, Lander, G.C, Wilson, I.A, Kelly, J.W. | | Deposit date: | 2023-09-11 | | Release date: | 2025-03-05 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The conformational landscape of human transthyretin revealed by cryo-EM.
Nat.Struct.Mol.Biol., 32, 2025
|
|
1MZ4
 
 | | Crystal Structure of Cytochrome c550 from Thermosynechococcus elongatus | | Descriptor: | BICARBONATE ION, GLYCEROL, HEME C, ... | | Authors: | Kerfeld, C.A, Sawaya, M.R, Bottin, H, Tran, K.T, Sugiura, M, Kirilovsky, D, Krogmann, D, Yeates, T.O, Boussac, A. | | Deposit date: | 2002-10-05 | | Release date: | 2003-09-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and EPR characterization of the soluble form of cytochrome c-550 and of the psbV2 gene product from the cyanobacterium Thermosynechococcus elongatus.
Plant Cell.Physiol., 44, 2003
|
|
8VCZ
 
 | |
8VD3
 
 | |
8VD1
 
 | | Crystal Structure of KAI2 from Oryza sativa with MPD | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Probable esterase D14L | | Authors: | Gilio, A.K, Shabek, N, Guercio, A.M, Pawlak, J. | | Deposit date: | 2023-12-14 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Structural insights into rice KAI2 receptor provide functional implications for perception and signal transduction.
J.Biol.Chem., 300, 2024
|
|
5F21
 
 | | human CD38 in complex with nanobody MU375 | | Descriptor: | ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1, GLYCEROL, SULFATE ION, ... | | Authors: | Hao, Q, Zhang, H. | | Deposit date: | 2015-12-01 | | Release date: | 2016-06-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Immuno-targeting the multifunctional CD38 using nanobody
Sci Rep, 6, 2016
|
|
5F1K
 
 | | human CD38 in complex with nanobody MU1053 | | Descriptor: | ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1, nanobody MU1053 | | Authors: | Zhang, H, Hao, Q. | | Deposit date: | 2015-11-30 | | Release date: | 2016-06-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Immuno-targeting the multifunctional CD38 using nanobody
Sci Rep, 6, 2016
|
|
5F1O
 
 | | human CD38 in complex with nanobody MU551 | | Descriptor: | ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1, nanobody MU551 | | Authors: | Zhang, H, Hao, Q. | | Deposit date: | 2015-11-30 | | Release date: | 2016-06-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Immuno-targeting the multifunctional CD38 using nanobody
Sci Rep, 6, 2016
|
|
8W0Q
 
 | | Pembrolizumab CDR-H3 Loop Mimic | | Descriptor: | Pembrolizumab CDR-H3 Loop Mimic | | Authors: | Feig, M, Roche, S.P. | | Deposit date: | 2024-02-14 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-31 | | Method: | SOLUTION NMR | | Cite: | De Novo Synthesis and Structural Elucidation of CDR-H3 Loop Mimics.
Acs Chem.Biol., 19, 2024
|
|
5DMR
 
 | |
4WL9
 
 | |
6W50
 
 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR METHYL ((10R,14S)- 14-(4-(3-CHLORO-2,6-DIFLUOROPHENYL)-6-OXO-3,6-DIHYDRO- 1(2H)-PYRIDINYL)-10-METHYL-9-OXO-8,16- DIAZATRICYCLO[13.3.1.0~2,7~]NONADECA-1(19),2,4,6,15,17- HEXAEN-5-YL)CARBAMATE | | Descriptor: | 1,2-ETHANEDIOL, Coagulation factor XI, SULFATE ION, ... | | Authors: | Sheriff, S. | | Deposit date: | 2020-03-12 | | Release date: | 2020-06-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of a High Affinity, Orally Bioavailable Macrocyclic FXIa Inhibitor with Antithrombotic Activity in Preclinical Species.
J.Med.Chem., 63, 2020
|
|
9EOQ
 
 | | Cryo-EM Structure of a 1033 Scaffold Base DNA Origami Nanostructure V4 and TBA | | Descriptor: | DNA (42-MER), DNA (5'-D(P*AP*TP*AP*TP*AP*GP*CP*GP*TP*GP*GP*AP*AP*GP*T)-3') | | Authors: | Ali, K, Georg, K, Volodymyr, M, Johanna, G, Maximilian, N.H, Lukas, K, Simone, C, Hendrik, D. | | Deposit date: | 2024-03-15 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Designing Rigid DNA Origami Templates for Molecular Visualization Using Cryo-EM.
Nano Lett., 24, 2024
|
|
5DMQ
 
 | |
6WQL
 
 | |
6G09
 
 | | Crystal Structure of a GH8 xylobiose complex from Teredinibacter turnerae | | Descriptor: | 1,2-ETHANEDIOL, Glycoside hydrolase family 8 domain protein, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Fowler, C.A, Davies, G.J, Walton, P.H. | | Deposit date: | 2018-03-16 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and function of a glycoside hydrolase family 8 endoxylanase from Teredinibacter turnerae.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6G0B
 
 | |
6G00
 
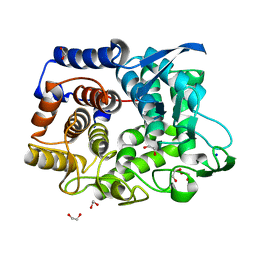 | | Crystal Structure of a GH8 xylanase from Teredinibacter turnerae | | Descriptor: | 1,2-ETHANEDIOL, Glycoside hydrolase family 8 domain protein, SODIUM ION | | Authors: | Fowler, C.A, Davies, G.J, Walton, P.H. | | Deposit date: | 2018-03-15 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and function of a glycoside hydrolase family 8 endoxylanase from Teredinibacter turnerae.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6G0N
 
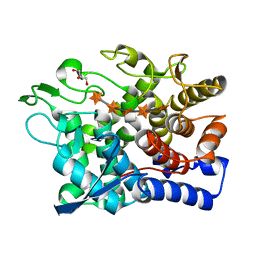 | | Crystal Structure of a GH8 catalytic mutant xylohexaose complex xylanase from Teredinibacter turnerae | | Descriptor: | GLYCEROL, Glycoside hydrolase family 8 domain protein, beta-D-xylopyranose, ... | | Authors: | Fowler, C.A, Davies, G.J, Walton, P.H. | | Deposit date: | 2018-03-19 | | Release date: | 2018-10-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and function of a glycoside hydrolase family 8 endoxylanase from Teredinibacter turnerae.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6G6E
 
 | |
7D2O
 
 | |
6GGJ
 
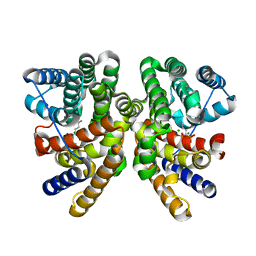 | | Crystal structure of CotB2 in complex with alendronate | | Descriptor: | 4-AMINO-1-HYDROXYBUTANE-1,1-DIYLDIPHOSPHONATE, Cyclooctat-9-en-7-ol synthase, MAGNESIUM ION | | Authors: | Driller, R, Janke, S, Fuchs, M, Warner, E, Mhashal, A.R, Major, D.T, Christmann, M, Brueck, T, Loll, B. | | Deposit date: | 2018-05-03 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Towards a comprehensive understanding of the structural dynamics of a bacterial diterpene synthase during catalysis.
Nat Commun, 9, 2018
|
|
