4UM1
 
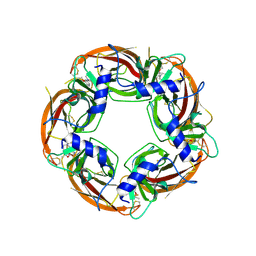 | | Engineered Ls-AChBP with alpha4-alpha4 binding pocket in complex with NS3573 | | Descriptor: | 1-(5-ethoxypyridin-3-yl)-1,4-diazepane, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINE-BINDING PROTEIN | | Authors: | Shahsavar, A, Kastrup, J.S, Balle, T, Gajhede, M. | | Deposit date: | 2014-05-14 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Achbp Engineered to Mimic the Alpha4-Alpha4 Binding Pocket in Alpha4Beta2 Nicotinic Acetylcholine Receptors Reveals Interface Specific Interactions Important for Binding and Activity
Mol.Pharmacol., 88, 2015
|
|
1YTQ
 
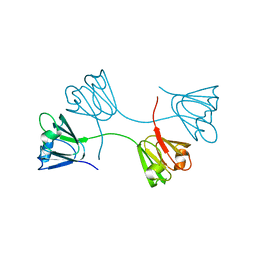 | |
4MZQ
 
 | |
6NB9
 
 | | Amyloid-Beta (20-34) with L-isoaspartate 23 | | Descriptor: | Amyloid-beta A4 protein | | Authors: | Sawaya, M.R, Warmack, R.A, Boyer, D.R, Zee, C.T, Richards, L.S, Cascio, D, Gonen, T, Clarke, S.G, Eisenberg, D.S. | | Deposit date: | 2018-12-06 | | Release date: | 2019-08-07 | | Last modified: | 2022-09-07 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.05 Å) | | Cite: | Structure of amyloid-beta (20-34) with Alzheimer's-associated isomerization at Asp23 reveals a distinct protofilament interface.
Nat Commun, 10, 2019
|
|
4PTZ
 
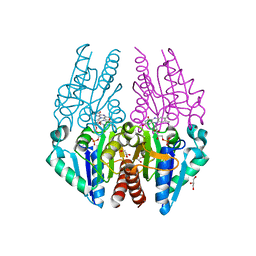 | | Crystal structure of the Escherichia coli alkanesulfonate FMN reductase SsuE in FMN-bound form | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN reductase SsuE, GLYCEROL, ... | | Authors: | Driggers, C.M, Ellis, H.R, Karplus, P.A. | | Deposit date: | 2014-03-11 | | Release date: | 2014-06-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9007 Å) | | Cite: | Crystal Structure of Escherichia coli SsuE: Defining a General Catalytic Cycle for FMN Reductases of the Flavodoxin-like Superfamily.
Biochemistry, 53, 2014
|
|
6OIZ
 
 | | Amyloid-Beta (20-34) wild type | | Descriptor: | Amyloid beta A4 protein | | Authors: | Sawaya, M.R, Warmack, R.A, Zee, C.T, Gonen, T, Clarke, S.G, Eisenberg, D.S. | | Deposit date: | 2019-04-10 | | Release date: | 2019-08-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.1 Å) | | Cite: | Structure of amyloid-beta (20-34) with Alzheimer's-associated isomerization at Asp23 reveals a distinct protofilament interface.
Nat Commun, 10, 2019
|
|
4PU0
 
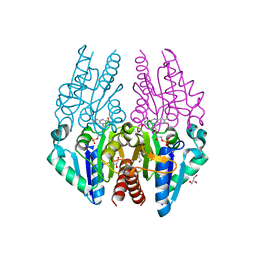 | | Crystal structure of the Escherichia coli alkanesulfonate FMN reductase SsuE in FMNH2-bound form | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN reductase SsuE, GLYCEROL, ... | | Authors: | Driggers, C.M, Ellis, H.R, Karplus, P.A. | | Deposit date: | 2014-03-11 | | Release date: | 2014-06-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3018 Å) | | Cite: | Crystal Structure of Escherichia coli SsuE: Defining a General Catalytic Cycle for FMN Reductases of the Flavodoxin-like Superfamily.
Biochemistry, 53, 2014
|
|
5IZ2
 
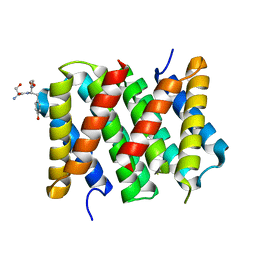 | | Crystal structure of the N. clavipes spidroin NTD at pH 6.5 | | Descriptor: | Major ampullate spidroin 1A, Major ampullate spidroin 1A (Partial C-terminus) | | Authors: | Atkison, J.H, Olsen, S.K. | | Deposit date: | 2016-03-24 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal Structure of the Nephila clavipes Major Ampullate Spidroin 1A N-terminal Domain Reveals Plasticity at the Dimer Interface.
J.Biol.Chem., 291, 2016
|
|
4PTY
 
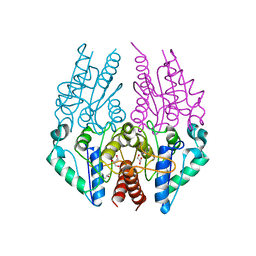 | |
7PJM
 
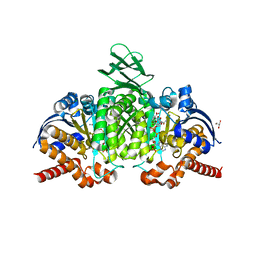 | | Crystal Structure of Ivosidenib-resistant IDH1 variant R132C S280F in complex with NADPH and Ca2+/2-Oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Reinbold, R, Rabe, P, Abboud, M.I, Schofield, C.J. | | Deposit date: | 2021-08-24 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Resistance to the isocitrate dehydrogenase 1 mutant inhibitor ivosidenib can be overcome by alternative dimer-interface binding inhibitors.
Nat Commun, 13, 2022
|
|
7PJN
 
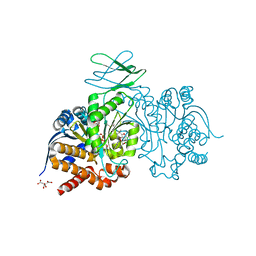 | | Crystal Structure of Ivosidenib-resistant IDH1 variant R132C S280F in complex with NADPH and inhibitor DS-1001B | | Descriptor: | (E)-3-(1-(5-(2-fluoropropan-2-yl)-3-(2,4,6-trichlorophenyl)isoxazole-4-carbonyl)-3-methyl-1H-indol-4-yl)acrylic acid, CITRIC ACID, GLYCEROL, ... | | Authors: | Reinbold, R, Rabe, P, Abboud, M.I, Schofield, C.J, Clifton, I.J. | | Deposit date: | 2021-08-24 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Resistance to the isocitrate dehydrogenase 1 mutant inhibitor ivosidenib can be overcome by alternative dimer-interface binding inhibitors.
Nat Commun, 13, 2022
|
|
6TB0
 
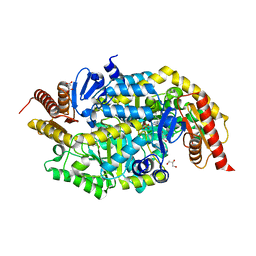 | | Crystal structure of thermostable omega transaminase 4-fold mutant from Pseudomonas jessenii | | Descriptor: | Aspartate aminotransferase family protein, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Capra, N, Rozeboom, H.J, Thunnissen, A.M.W.H, Janssen, D.B. | | Deposit date: | 2019-10-31 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Robust omega-Transaminases by Computational Stabilization of the Subunit Interface.
Acs Catalysis, 10, 2020
|
|
6TB1
 
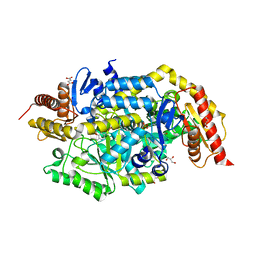 | | Crystal structure of thermostable omega transaminase 6-fold mutant from Pseudomonas jessenii | | Descriptor: | Aspartate aminotransferase family protein, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Capra, N, Rozeboom, H.J, Thunnissen, A.M.W.H, Janssen, D.B. | | Deposit date: | 2019-10-31 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Robust omega-Transaminases by Computational Stabilization of the Subunit Interface.
Acs Catalysis, 10, 2020
|
|
6WLE
 
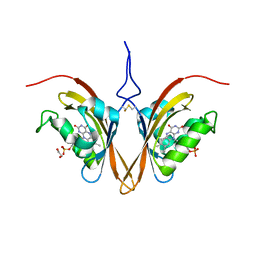 | | Crystal structure of the Zeitlupe light-state mimic G46A | | Descriptor: | 1,2-ETHANEDIOL, Adagio protein 1, FLAVIN MONONUCLEOTIDE | | Authors: | Zoltowski, B, Green, R. | | Deposit date: | 2020-04-20 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Steric and Electronic Interactions at Gln154 in ZEITLUPE Induce Reorganization of the LOV Domain Dimer Interface.
Biochemistry, 60, 2021
|
|
6WLP
 
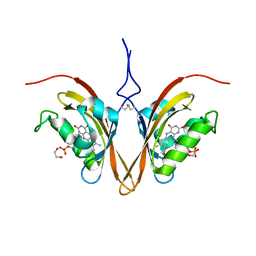 | | Crystal Structure of the ZTL light-state mimic G46S | | Descriptor: | 1,2-ETHANEDIOL, Adagio protein 1, FLAVIN MONONUCLEOTIDE | | Authors: | Zoltowski, B, Green, R. | | Deposit date: | 2020-04-20 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Steric and Electronic Interactions at Gln154 in ZEITLUPE Induce Reorganization of the LOV Domain Dimer Interface.
Biochemistry, 60, 2021
|
|
4UM3
 
 | | Engineered Ls-AChBP with alpha4-alpha4 binding pocket in complex with NS3920 | | Descriptor: | 1-(6-bromopyridin-3-yl)-1,4-diazepane, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINE BINDING PROTEIN, ... | | Authors: | Shahsavar, A, Kastrup, J.S, Balle, T, Gajhede, M. | | Deposit date: | 2014-05-14 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Achbp Engineered to Mimic the Alpha4-Alpha4 Binding Pocket in Alpha4Beta2 Nicotinic Acetylcholine Receptors Reveals Interface Specific Interactions Important for Binding and Activity
Mol.Pharmacol., 88, 2015
|
|
6RWH
 
 | | Fragment AZ-007 binding at a primary and secondary binding site of the the p53pT387/14-3-3 sigma complex | | Descriptor: | 14-3-3 protein sigma, 5-(1~{H}-imidazol-5-yl)-4-phenyl-thiophene-2-carboximidamide, CALCIUM ION, ... | | Authors: | Leysen, S, Guillory, X, Wolter, M, Genet, S, Somsen, B, Patel, J, Castaldi, P, Ottmann, C. | | Deposit date: | 2019-06-05 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
5LVE
 
 | |
6S9Q
 
 | | Fragment AZ-004 binding at a primary and secondary site in a p53pT387/14-3-3 complex | | Descriptor: | 14-3-3 protein sigma, 4-methyl-5-phenyl-thiophene-2-carboximidamide, CALCIUM ION, ... | | Authors: | Leysen, S, Guillory, X, Wolter, M, Genet, S, Somsen, B, Patel, J, Castaldi, P, Ottmann, C. | | Deposit date: | 2019-07-15 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
1MUD
 
 | | CATALYTIC DOMAIN OF MUTY FROM ESCHERICHIA COLI, D138N MUTANT COMPLEXED TO ADENINE | | Descriptor: | ADENINE, Adenine DNA glycosylase, GLYCEROL, ... | | Authors: | Guan, Y, Tainer, J.A. | | Deposit date: | 1998-08-20 | | Release date: | 1999-09-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | MutY catalytic core, mutant and bound adenine structures define specificity for DNA repair enzyme superfamily.
Nat.Struct.Biol., 5, 1998
|
|
3FG6
 
 | | Structure of the C-terminus of Adseverin | | Descriptor: | Adseverin, CALCIUM ION | | Authors: | Robinson, R.C. | | Deposit date: | 2008-12-05 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of the C-terminus of adseverin reveals the actin-binding interface.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4OEJ
 
 | | Structure of membrane binding protein pleurotolysin B from Pleurotus ostreatus | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Dunstone, M.A, Caradoc-Davies, T.T, Whisstock, J.C, Law, R.H.P. | | Deposit date: | 2014-01-13 | | Release date: | 2015-02-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational Changes during Pore Formation by the Perforin-Related Protein Pleurotolysin.
Plos Biol., 13, 2015
|
|
5KEH
 
 | |
5KF3
 
 | |
4OEB
 
 | | Structure of membrane binding protein pleurotolysin A from Pleurotus ostreatus | | Descriptor: | Pleurotolysin A, SULFATE ION | | Authors: | Dunstone, M.A, Caradoc-Davies, T.T, Whisstock, J.C, Law, R.H.P. | | Deposit date: | 2014-01-12 | | Release date: | 2015-02-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Conformational Changes during Pore Formation by the Perforin-Related Protein Pleurotolysin.
Plos Biol., 13, 2015
|
|
