7YZD
 
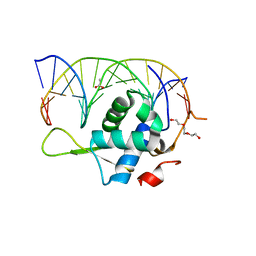 | |
7YZG
 
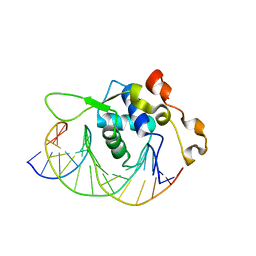 | | Crystal structure of the Xenopus FoxH1 bound to the TGTGGATT site | | Descriptor: | DNA (5'-D(*CP*AP*GP*AP*TP*TP*GP*TP*GP*GP*AP*TP*TP*GP*AP*G)-3'), DNA (5'-D(*CP*TP*CP*AP*AP*TP*CP*CP*AP*CP*AP*AP*TP*CP*TP*G)-3'), Forkhead box protein H1 | | Authors: | Pluta, R, Macias, M.J. | | Deposit date: | 2022-02-19 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Molecular basis for DNA recognition by the maternal pioneer transcription factor FoxH1.
Nat Commun, 13, 2022
|
|
7YZC
 
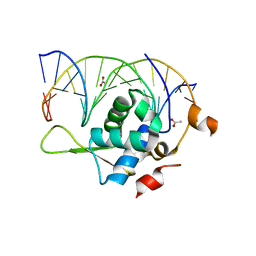 | | Crystal structure of the zebrafish FoxH1 bound to the TGTTTATT site | | Descriptor: | ACETATE ION, DNA (5'-D(*AP*GP*AP*TP*TP*GP*TP*TP*TP*AP*TP*TP*GP*AP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*CP*AP*AP*TP*AP*AP*AP*CP*AP*AP*TP*CP*T)-3'), ... | | Authors: | Pluta, R, Macias, M.J. | | Deposit date: | 2022-02-19 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Molecular basis for DNA recognition by the maternal pioneer transcription factor FoxH1.
Nat Commun, 13, 2022
|
|
7YZE
 
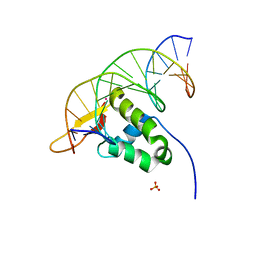 | |
7PDV
 
 | |
7YZF
 
 | |
7YZR
 
 | | 50 mM Rb+ soak of beryllium fluoride inhibited Na+,K+-ATPase, E2-BeFx (rigid body model) | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, FXYD domain-containing ion transport regulator, MAGNESIUM ION, ... | | Authors: | Fruergaard, M.U, Dach, I, Andersen, J.L, Ozol, M, Shasavar, A, Quistgaard, E.M, Poulsen, H, Fedosova, N.U, Nissen, P. | | Deposit date: | 2022-02-21 | | Release date: | 2022-11-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (6.92 Å) | | Cite: | The Na + ,K + -ATPase in complex with beryllium fluoride mimics an ATPase phosphorylated state.
J.Biol.Chem., 298, 2022
|
|
4ZBI
 
 | | Mcl-1 complexed with small molecules | | Descriptor: | 1-[3-(naphthalen-1-yloxy)propyl]-5,6-dihydro-4H-pyrrolo[3,2,1-ij]quinoline-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Zhao, B. | | Deposit date: | 2015-04-14 | | Release date: | 2015-04-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of tricyclic indoles that potently inhibit mcl-1 using fragment-based methods and structure-based design.
J.Med.Chem., 58, 2015
|
|
7Z04
 
 | | 10 mM Rb+ soak of beryllium fluoride inhibited Na+,K+-ATPase, E2-BeFx (rigid body model) | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, FXYD domain-containing ion transport regulator, MAGNESIUM ION, ... | | Authors: | Fruergaard, M.U, Dach, I, Andersen, J.L, Ozol, M, Shasavar, A, Quistgaard, E.M, Poulsen, H, Fedosova, N.U, Nissen, P. | | Deposit date: | 2022-02-22 | | Release date: | 2022-11-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (7.5 Å) | | Cite: | The Na + ,K + -ATPase in complex with beryllium fluoride mimics an ATPase phosphorylated state.
J.Biol.Chem., 298, 2022
|
|
6MYX
 
 | | EM structure of Bacillus subtilis ribonucleotide reductase inhibited double-helical filament of NrdE alpha subunit with dATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Ribonucleoside-diphosphate reductase | | Authors: | Thomas, W.C, Bacik, J.P, Chen, J.Z, Ando, N. | | Deposit date: | 2018-11-02 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Convergent allostery in ribonucleotide reductase.
Nat Commun, 10, 2019
|
|
7YLA
 
 | | Cryo-EM structure of 50S-HflX complex | | Descriptor: | 50S ribosomal protein L11, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Damu, W, Ning, G. | | Deposit date: | 2022-07-25 | | Release date: | 2023-01-04 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Cryo-EM Structure of the 50S-HflX Complex Reveals a Novel Mechanism of Antibiotic Resistance in E. coli
Biorxiv, 2022
|
|
4ZMA
 
 | | Crystal Structure of a FVIIa-Trypsin Chimera (ST) in Complex with Soluble Tissue Factor | | Descriptor: | CACODYLATE ION, CALCIUM ION, Coagulation factor VII, ... | | Authors: | Sorensen, A.B, Svensson, L.A, Gandhi, P.S. | | Deposit date: | 2015-05-02 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular Basis of Enhanced Activity in Factor VIIa-Trypsin Variants Conveys Insights into Tissue Factor-mediated Allosteric Regulation of Factor VIIa Activity.
J.Biol.Chem., 291, 2016
|
|
2Y6L
 
 | | Xylopentaose binding X-2 engineered mutated CBM4-2 Carbohydrate Binding Module from a Thermostable Rhodothermus marinus Xylanase | | Descriptor: | CALCIUM ION, XYLANASE, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | von Schantz, L, Hakansson, M, Logan, D.T, Walse, B, Osterlin, J, Nordberg-Karlsson, E, Ohlin, M. | | Deposit date: | 2011-01-24 | | Release date: | 2012-03-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structural basis for carbohydrate-binding specificity--a comparative assessment of two engineered carbohydrate-binding modules.
Glycobiology, 22, 2012
|
|
7P33
 
 | | Epstein-Barr virus encoded Bcl-2 homolog BHRF-1 in complex with Bid BH3 peptide | | Descriptor: | 1,2-ETHANEDIOL, Apoptosis regulator BHRF1, BH3-interacting domain death agonist p15, ... | | Authors: | Suraweera, C.D, Hinds, M.G, Kvansakul, M. | | Deposit date: | 2021-07-07 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.78542733 Å) | | Cite: | Crystal Structures of Epstein-Barr Virus Bcl-2 Homolog BHRF1 Bound to Bid and Puma BH3 Motif Peptides.
Viruses, 14, 2022
|
|
4ZRJ
 
 | | Structure of Merlin-FERM and CTD | | Descriptor: | GLYCEROL, Merlin | | Authors: | Lin, Z, Li, F, Long, J, Shen, Y. | | Deposit date: | 2015-05-12 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Angiomotin binding-induced activation of Merlin/NF2 in the Hippo pathway
Cell Res., 25, 2015
|
|
4Z6A
 
 | | Crystal Structure of a FVIIa-Trypsin Chimera (YT) in Complex with Soluble Tissue Factor | | Descriptor: | CALCIUM ION, CITRIC ACID, Coagulation factor VII, ... | | Authors: | Sorensen, A.B, Svensson, L.A, Gandhi, P.S. | | Deposit date: | 2015-04-04 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Molecular Basis of Enhanced Activity in Factor VIIa-Trypsin Variants Conveys Insights into Tissue Factor-mediated Allosteric Regulation of Factor VIIa Activity.
J.Biol.Chem., 291, 2016
|
|
4Z7I
 
 | | Crystal structure of insulin regulated aminopeptidase in complex with ligand | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DG025 transition-state analogue enzyme inhibitor, ... | | Authors: | Mpakali, A, Saridakis, E, Harlos, K, Zhao, Y, Stratikos, E. | | Deposit date: | 2015-04-07 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Crystal Structure of Insulin-Regulated Aminopeptidase with Bound Substrate Analogue Provides Insight on Antigenic Epitope Precursor Recognition and Processing.
J Immunol., 195, 2015
|
|
6N69
 
 | | rat hPGDS complexed with a quinoline | | Descriptor: | GLUTATHIONE, Hematopoietic prostaglandin D synthase, quinoline-3-carbonitrile | | Authors: | Shewchuk, L.M, Cleasby, A. | | Deposit date: | 2018-11-26 | | Release date: | 2019-03-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The discovery of quinoline-3-carboxamides as hematopoietic prostaglandin D synthase (H-PGDS) inhibitors.
Bioorg. Med. Chem., 27, 2019
|
|
4ZHA
 
 | | Factor Xa complex with GTC000102 | | Descriptor: | 4-[(3S)-3-({[(E)-2-(5-chlorothiophen-2-yl)ethenyl]sulfonyl}amino)-2-oxo-2,3-dihydro-1H-pyrrol-1-yl]-3-fluoro-N-methylbenzamide, CALCIUM ION, Coagulation factor X, ... | | Authors: | Convery, M.A. | | Deposit date: | 2015-04-24 | | Release date: | 2016-01-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Factor Xa complex with GTC000102
To Be Published
|
|
4ZII
 
 | | Crystal Structure of core/latch dimer of BaxI66A in complex with BidBH3 | | Descriptor: | 1,2-ETHANEDIOL, Apoptosis regulator BAX, BH3-interacting domain death agonist | | Authors: | Czabotar, P.E, Robin, A.Y, Krishna Kumar, K, Westphal, D, Wardak, A.Z, Thompson, G.V, Dewson, G, Colman, P.M. | | Deposit date: | 2015-04-28 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.191 Å) | | Cite: | Crystal structure of Bax bound to the BH3 peptide of Bim identifies important contacts for interaction.
Cell Death Dis, 6, 2015
|
|
7YF5
 
 | |
7YAN
 
 | |
6MW3
 
 | | EM structure of Bacillus subtilis ribonucleotide reductase inhibited filament composed of NrdE alpha subunit and NrdF beta subunit with dATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Ribonucleoside-diphosphate reductase, Ribonucleoside-diphosphate reductase NrdF beta subunit | | Authors: | Thomas, W.C, Bacik, J.P, Kaelber, J.T, Ando, N. | | Deposit date: | 2018-10-29 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.65 Å) | | Cite: | Convergent allostery in ribonucleotide reductase.
Nat Commun, 10, 2019
|
|
4ZIG
 
 | | Crystal Structure of core/latch dimer of Bax in complex with BidBH3mini | | Descriptor: | Apoptosis regulator BAX, BH3-interacting domain death agonist | | Authors: | Robin, A.Y, Krishna Kumar, K, Westphal, D, Wardak, A.Z, Thompson, G.V, Dewson, G, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2015-04-28 | | Release date: | 2015-07-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Bax bound to the BH3 peptide of Bim identifies important contacts for interaction.
Cell Death Dis, 6, 2015
|
|
7P9W
 
 | | Epstein-Barr virus encoded apoptosis regulator BHRF1 in complex with Puma BH3 | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, AMMONIUM ION, ... | | Authors: | Suraweera, C.D, Hinds, M.G, Kvansakul, M. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.00010061 Å) | | Cite: | Crystal Structures of Epstein-Barr Virus Bcl-2 Homolog BHRF1 Bound to Bid and Puma BH3 Motif Peptides.
Viruses, 14, 2022
|
|
