1MIW
 
 | | Crystal structure of Bacillus stearothermophilus CCA-adding enzyme in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, tRNA CCA-adding enzyme | | Authors: | Li, F, Xiong, Y, Wang, J, Cho, H.D, Weiner, A.M, Steitz, T.A. | | Deposit date: | 2002-08-23 | | Release date: | 2002-12-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of the Bacillus stearothermophilus CCA-adding enzyme and
its complexes with ATP or CTP
Cell(Cambridge,Mass.), 111, 2002
|
|
1MJF
 
 | |
4KOB
 
 | |
4DIX
 
 | |
4KQS
 
 | | Crystal Structure of Farnesyl Pyrophosphate Synthase Mutant (Y204A) Complexed with Mg, Risedronate and Isopentenyl Pyrophosphate | | Descriptor: | 1-HYDROXY-2-(3-PYRIDINYL)ETHYLIDENE BIS-PHOSPHONIC ACID, 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Farnesyl pyrophosphate synthase, ... | | Authors: | Barnett, B.L, Tsoumpra, M.K, Muniz, J.R.C. | | Deposit date: | 2013-05-15 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal Structure of Farnesyl Pyrophosphate Synthase Mutant (Y204A) Complexed with Mg, Risedronate and Isopentenyl Pyrophosphate
To be Published
|
|
1MPX
 
 | | ALPHA-AMINO ACID ESTER HYDROLASE LABELED WITH SELENOMETHIONINE | | Descriptor: | CALCIUM ION, GLYCEROL, alpha-amino acid ester hydrolase | | Authors: | Barends, T.R.M, Polderman-Tijmes, J.J, Jekel, P.A, Hensgens, C.M.H, de Vries, E.J, Janssen, D.B, Dijkstra, B.W. | | Deposit date: | 2002-09-13 | | Release date: | 2003-04-15 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The sequence and crystal structure of the alpha-amino acid ester hydrolase from Xanthomonas citri define a new family of beta-lactam antibiotic acylases.
J.Biol.Chem., 278, 2003
|
|
1MKZ
 
 | | Crystal structure of MoaB protein at 1.6 A resolution. | | Descriptor: | ACETIC ACID, Molybdenum cofactor biosynthesis protein B, SULFATE ION | | Authors: | Sanishvili, R, Skarina, T, Joachimiak, A, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-08-29 | | Release date: | 2003-04-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of Escherichia coli MoaB suggests a probable role in molybdenum cofactor synthesis.
J.Biol.Chem., 279, 2004
|
|
1MO2
 
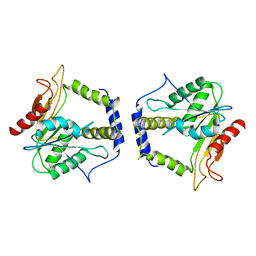 | | Thioesterase Domain from 6-Deoxyerythronolide Synthase (DEBS TE), pH 8.5 | | Descriptor: | Erythronolide synthase, modules 5 and 6 | | Authors: | Tsai, S.-C, Lu, H, Cane, D.E, Khosla, C, Stroud, R.M. | | Deposit date: | 2002-09-05 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Insights into channel architecture and substrate specificity from crystal structures of two macrocycle-forming thioesterases of modular polyketide synthases
Biochemistry, 41, 2002
|
|
8JYD
 
 | |
8JZ6
 
 | |
1M33
 
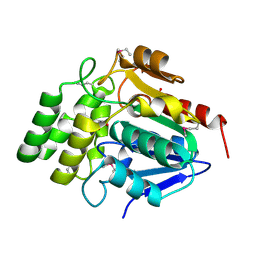 | | Crystal Structure of BioH at 1.7 A | | Descriptor: | 1,2-ETHANEDIOL, 3-HYDROXY-PROPANOIC ACID, BioH protein | | Authors: | Sanishvili, R, Savchenko, A, Skarina, T, Edwards, A, Joachimiak, A, Yakunin, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-06-26 | | Release date: | 2003-01-21 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Integrating structure, bioinformatics, and enzymology to discover function: BioH, a new carboxylesterase from Escherichia coli.
J.Biol.Chem., 278, 2003
|
|
8K0O
 
 | |
4DHO
 
 | | Small-molecule inhibitors of 14-3-3 protein-protein interactions from virtual screening | | Descriptor: | (2-{2-[(3-methoxyphenyl)amino]-2-oxoethoxy}phenyl)phosphonic acid, 14-3-3 protein sigma, CHLORIDE ION, ... | | Authors: | Thiel, P, Roeglin, L, Kohlbacher, O, Ottmann, C. | | Deposit date: | 2012-01-30 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Virtual screening and experimental validation reveal novel small-molecule inhibitors of 14-3-3 protein-protein interactions.
Chem.Commun.(Camb.), 49, 2013
|
|
4DHU
 
 | | Small-molecule inhibitors of 14-3-3 protein-protein interactions from virtual screening | | Descriptor: | (2-{2-[(2,3-dichlorophenyl)amino]-2-oxoethoxy}phenyl)phosphonic acid, 14-3-3 PROTEIN SIGMA, CHLORIDE ION, ... | | Authors: | Thiel, P, Roeglin, L, Kohlbacher, O, Ottmann, C. | | Deposit date: | 2012-01-30 | | Release date: | 2013-07-31 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Virtual screening and experimental validation reveal novel small-molecule inhibitors of 14-3-3 protein-protein interactions.
Chem.Commun.(Camb.), 49, 2013
|
|
8KA0
 
 | | Crystal structure of Vibrio vulnificus RID-dependent transforming NADase domain (RDTND)/calmodulin-binding domain of Rho inactivation domain (RID-CBD) complexed with Ca2+-bound calmodulin and a nicotinamide adenine dinucleotide (NAD+) | | Descriptor: | CALCIUM ION, Calmodulin-2, GLYCEROL, ... | | Authors: | Lee, Y, Choi, S, Hwang, J, Kim, M.H. | | Deposit date: | 2023-08-02 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Dissemination of pathogenic bacteria is reinforced by a MARTX toxin effector duet.
Nat Commun, 15, 2024
|
|
4KU4
 
 | |
8KA2
 
 | |
8KG4
 
 | | Crystal Structure of M- and C-Domains of the shaft pilin LrpA from Ligilactobacillus ruminis - orthorhombic form | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, IODIDE ION, ... | | Authors: | Prajapati, A, Palva, A, von Ossowski, I, Krishnan, V. | | Deposit date: | 2023-08-17 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The crystal structure of the N-terminal domain of the backbone pilin LrpA reveals a new closure-and-twist motion for assembling dynamic pili in Ligilactobacillus ruminis.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
4L2D
 
 | |
8K9Z
 
 | | Crystal structure of Vibrio vulnificus RID-dependent transforming NADase domain (RDTND)/calmodulin-binding domain of Rho inactivation domain (RID-CBD) complexed with Ca2+-bound calmodulin | | Descriptor: | CALCIUM ION, Calmodulin-2, RDTND-RID CBD | | Authors: | Lee, Y, Choi, S, Hwang, J, Kim, M.H. | | Deposit date: | 2023-08-02 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Dissemination of pathogenic bacteria is reinforced by a MARTX toxin effector duet.
Nat Commun, 15, 2024
|
|
8K4D
 
 | | Structure of the SA2/Scc1/CENP_U complex | | Descriptor: | 64-kDa C-terminal product, CENP-U, Cohesin subunit SA-2 | | Authors: | Liu, M.J, He, X.J. | | Deposit date: | 2023-07-18 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | The CENP-O complex links inner kinetochore to centromere cohesion
To Be Published
|
|
8KB2
 
 | | Crystal Structure of M- and C-Domains of the shaft pilin LrpA from Ligilactobacillus ruminis - iodide derivative | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Prajapati, A, Palva, A, von Ossowski, I, Krishnan, V. | | Deposit date: | 2023-08-03 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | The crystal structure of the N-terminal domain of the backbone pilin LrpA reveals a new closure-and-twist motion for assembling dynamic pili in Ligilactobacillus ruminis.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8KA1
 
 | | Crystal structure of Vibrio vulnificus RID-dependent transforming NADase domain (RDTND)/calmodulin-binding domain of Rho inactivation domain (RID-CBD) complexed with Ca2+-free calmodulin | | Descriptor: | Calmodulin-2, MAGNESIUM ION, RDTND-RID CBD | | Authors: | Lee, Y, Choi, S, Hwang, J, Kim, M.H. | | Deposit date: | 2023-08-02 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Dissemination of pathogenic bacteria is reinforced by a MARTX toxin effector duet.
Nat Commun, 15, 2024
|
|
1MFV
 
 | | Probing the role of a mobile loop in human slaivary amylase: Structural studies on the loop-deleted enzyme | | Descriptor: | 4-amino-4,6-dideoxy-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 4-amino-4,6-dideoxy-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, 5-HYDROXYMETHYL-CHONDURITOL, ... | | Authors: | Ramasubbu, N, Ragunath, C, Mishra, P.J. | | Deposit date: | 2002-08-13 | | Release date: | 2002-11-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the role of a mobile loop in substrate binding and enzyme activity of
human salivary amylase.
J.Mol.Biol., 325, 2003
|
|
8KCL
 
 | |
