7CK5
 
 | | Solution structure of 28 amino acid polypeptide (354-381) in Plantago asiatica mosaic virus replicase bound to SDS micelle | | Descriptor: | PlAMV replicase peptide from RNA-dependent RNA polymerase | | Authors: | Komatsu, K, Sasaki, N, Yoshida, T, Suzuki, K, Masujima, Y, Hashimoto, M, Watanabe, S, Tochio, N, Kigawa, T, Yamaji, Y, Oshima, K, Namba, S, Nelson, R, Arie, T. | | Deposit date: | 2020-07-15 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Identification of a Proline-Kinked Amphipathic alpha-Helix Downstream from the Methyltransferase Domain of a Potexvirus Replicase and Its Role in Virus Replication and Perinuclear Complex Formation.
J.Virol., 95, 2021
|
|
7LRQ
 
 | | Crystal structure of human SFPQ/NONO heterodimer, conserved DBHS region | | Descriptor: | CHLORIDE ION, Non-POU domain-containing octamer-binding protein, Splicing factor, ... | | Authors: | Marshall, A.C, Bond, C.S, Mohnen, I. | | Deposit date: | 2021-02-17 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Paraspeckle subnuclear bodies depend on dynamic heterodimerisation of DBHS RNA-binding proteins via their structured domains.
J.Biol.Chem., 298, 2022
|
|
5K9D
 
 | | Crystal structure of human dihydroorotate dehydrogenase at 1.7 A resolution | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Lewis, T.A, Sykes, D.B, Law, J.M, Munoz, B, Scadden, D.T, Rustiguel, J.K, Nonato, M.C, Schreiber, S.L. | | Deposit date: | 2016-05-31 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Development of ML390: A Human DHODH Inhibitor That Induces Differentiation in Acute Myeloid Leukemia.
ACS Med Chem Lett, 7, 2016
|
|
5K9C
 
 | | Crystal structure of human dihydroorotate dehydrogenase with ML390 | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Lewis, T.A, Sykes, D.B, Law, J.M, Munoz, B, Scadden, D.T, Rustiguel, J.K, Nonato, M.C, Schreiber, S.L. | | Deposit date: | 2016-05-31 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Development of ML390: A Human DHODH Inhibitor That Induces Differentiation in Acute Myeloid Leukemia.
ACS Med Chem Lett, 7, 2016
|
|
8I4J
 
 | |
8I4K
 
 | | Structure of Azami Red1.0, a red fluorescent protein engineered from Azami Green | | Descriptor: | Azami Red1.0, CALCIUM ION | | Authors: | Otsubo, S, Takekawa, N, Imamura, H, Imada, K. | | Deposit date: | 2023-01-19 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Red fluorescent proteins engineered from green fluorescent proteins.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8CUY
 
 | | ACP1-KS-AT domains of mycobacterial Pks13 | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Polyketide synthase PKS13, UNKNOWN LIGAND | | Authors: | Kim, S.K, Dickinson, M.S, Finer-Moore, J.S, Rosenberg, O.S, Stroud, R.M. | | Deposit date: | 2022-05-17 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structure and dynamics of the essential endogenous mycobacterial polyketide synthase Pks13.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8CV0
 
 | | KS-AT domains of mycobacterial Pks13 with outward AT conformation | | Descriptor: | Polyketide synthase PKS13, UNKNOWN LIGAND | | Authors: | Kim, S.K, Dickinson, M.S, Finer-Moore, J.S, Rosenberg, O.S, Stroud, R.M. | | Deposit date: | 2022-05-17 | | Release date: | 2023-02-15 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure and dynamics of the essential endogenous mycobacterial polyketide synthase Pks13.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8CUZ
 
 | | KS-AT domains of mycobacterial Pks13 with inward AT conformation | | Descriptor: | Polyketide synthase PKS13, UNKNOWN LIGAND | | Authors: | Kim, S.K, Dickinson, M.S, Finer-Moore, J.S, Rosenberg, O.S, Stroud, R.M. | | Deposit date: | 2022-05-17 | | Release date: | 2023-02-15 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure and dynamics of the essential endogenous mycobacterial polyketide synthase Pks13.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8CV1
 
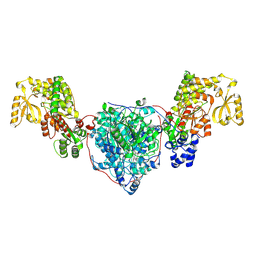 | | ACP1-KS-AT domains of mycobacterial Pks13 | | Descriptor: | Polyketide synthase PKS13, UNKNOWN LIGAND | | Authors: | Kim, S.K, Dickinson, M.S, Finer-Moore, J.S, Rosenberg, O.S, Stroud, R.M. | | Deposit date: | 2022-05-17 | | Release date: | 2023-02-15 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure and dynamics of the essential endogenous mycobacterial polyketide synthase Pks13.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5WD6
 
 | | bovine salivary protein form 30b | | Descriptor: | CALCIUM ION, Short palate, lung and nasal epithelium carcinoma-associated protein 2B | | Authors: | Zhang, H, Arcus, V.L. | | Deposit date: | 2017-07-04 | | Release date: | 2018-07-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The three dimensional structure of Bovine Salivary Protein 30b (BSP30b) and its interaction with specific rumen bacteria.
Plos One, 14, 2019
|
|
7UGS
 
 | |
7XM9
 
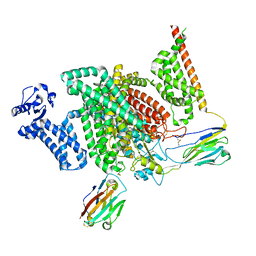 | | Cryo-EM structure of human NaV1.7/beta1/beta2-XEN907 | | Descriptor: | (7~{R})-1'-pentylspiro[6~{H}-furo[3,2-f][1,3]benzodioxole-7,3'-indole]-2'-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | zhang, J.T, Jiang, D.H. | | Deposit date: | 2022-04-25 | | Release date: | 2022-11-30 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structural basis for Na V 1.7 inhibition by pore blockers.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7XMF
 
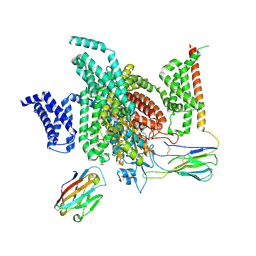 | | Cryo-EM structure of human NaV1.7/beta1/beta2-Nav1.7-IN2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[[4-[3-(4-fluoranyl-2-methyl-phenoxy)azetidin-1-yl]pyrimidin-2-yl]amino]-~{N}-methyl-benzamide, ... | | Authors: | Zhang, J.T, Jiang, D.H. | | Deposit date: | 2022-04-25 | | Release date: | 2022-11-30 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural basis for Na V 1.7 inhibition by pore blockers.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8HSI
 
 | | Cryo-EM structure of human TMEM87A, PE-bound | | Descriptor: | (1S)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(octadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Han, A, Kim, H.M. | | Deposit date: | 2022-12-19 | | Release date: | 2023-12-27 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | GolpHCat (TMEM87A), a unique voltage-dependent cation channel in Golgi apparatus, contributes to Golgi-pH maintenance and hippocampus-dependent memory.
Nat Commun, 15, 2024
|
|
7QLO
 
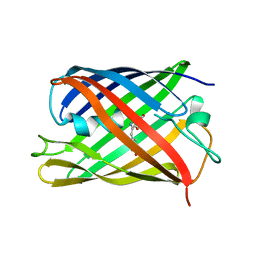 | | rsKiiro pump dump probe structure by TR-SFX | | Descriptor: | rsKiiro | | Authors: | van Thor, J.J. | | Deposit date: | 2021-12-20 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Optical control of ultrafast structural dynamics in a fluorescent protein.
Nat.Chem., 15, 2023
|
|
6PFU
 
 | |
6PFR
 
 | |
6PFS
 
 | |
6PFT
 
 | |
6OA8
 
 | | Superfolder Green Fluorescent Protein with 4-cyano-L-phenylalanine at the chromophore (position 66) | | Descriptor: | 1,2-ETHANEDIOL, Green fluorescent protein, SODIUM ION, ... | | Authors: | Piacentini, J, Olenginski, G.M, Brewer, S.H, Phillips-Piro, C.M. | | Deposit date: | 2019-03-15 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structural and spectrophotometric investigation of two unnatural amino-acid altered chromophores in the superfolder green fluorescent protein
Acta Crystallogr.,Sect.D, 2021
|
|
9DK8
 
 | | TRIF TIR Filament Cryo-EM Structure | | Descriptor: | TIR domain-containing adapter molecule 1 | | Authors: | Manik, M.K, Xiao, L, Wu, H. | | Deposit date: | 2024-09-08 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for TIR domain-mediated innate immune signaling by Toll-like receptor adaptors TRIF and TRAM.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
6MGH
 
 | | X-ray structure of monomeric near-infrared fluorescent protein miRFP670nano | | Descriptor: | 3-[2-[(~{Z})-[5-[(~{Z})-[(3~{S},4~{R})-3-ethenyl-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-pyrrol-2-ylidene]methyl]-5-[(~{Z})-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Pletnev, S. | | Deposit date: | 2018-09-13 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Smallest near-infrared fluorescent protein evolved from cyanobacteriochrome as versatile tag for spectral multiplexing.
Nat Commun, 10, 2019
|
|
4FL6
 
 | | Crystal structure of the complex of the 3-MBT repeat domain of L3MBTL3 and UNC1215 | | Descriptor: | Lethal(3)malignant brain tumor-like protein 3, UNKNOWN ATOM OR ION, [2-(phenylamino)benzene-1,4-diyl]bis{[4-(pyrrolidin-1-yl)piperidin-1-yl]methanone} | | Authors: | Zhong, N, Tempel, W, Ravichandran, M, Dong, A, Ingerman, L.A, Graslund, S, Frye, S.V, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-06-14 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery of a chemical probe for the L3MBTL3 methyllysine reader domain.
Nat. Chem. Biol., 9, 2013
|
|
6O1T
 
 | | BOVINE SALIVARY PROTEIN FORM 30B WITH OLEIC ACID | | Descriptor: | CALCIUM ION, OLEIC ACID, Short palate, ... | | Authors: | Zhang, H, Arcus, V.L. | | Deposit date: | 2019-02-21 | | Release date: | 2019-03-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The three dimensional structure of Bovine Salivary Protein 30b (BSP30b) and its interaction with specific rumen bacteria.
Plos One, 14, 2019
|
|
