1XRY
 
 | | Crystal structure of Aeromonas proteolytica aminopeptidase in complex with bestatin | | Descriptor: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, Bacterial leucyl aminopeptidase, ZINC ION | | Authors: | Gilboa, R, Rondeau, J.-M, Blumberg, S, Tarnus, C, Shoham, G. | | Deposit date: | 2004-10-17 | | Release date: | 2005-09-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Interactions of Streptomyces griseus Aminopeptidase and Aeromonas proteolytica Aminopeptidase with Bestatin. Structural analysis of homologous enzymes with different binding modes.
To be Published
|
|
1JBW
 
 | | FPGS-AMPPCP-folate complex | | Descriptor: | 5,10-METHYLENE-6-HYDROFOLIC ACID, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, FOLYLPOLYGLUTAMATE SYNTHASE, ... | | Authors: | Sun, X, Cross, J.A, Bognar, A.L, Baker, E.N, Smith, C.A. | | Deposit date: | 2001-06-06 | | Release date: | 2001-09-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Folate-binding triggers the activation of folylpolyglutamate synthetase.
J.Mol.Biol., 310, 2001
|
|
1JBV
 
 | | FPGS-AMPPCP complex | | Descriptor: | FOLYLPOLYGLUTAMATE SYNTHASE, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Sun, X, Cross, J.A, Bognar, A.L, Baker, E.N, Smith, C.A. | | Deposit date: | 2001-06-06 | | Release date: | 2001-09-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Folate-binding triggers the activation of folylpolyglutamate synthetase.
J.Mol.Biol., 310, 2001
|
|
3B3W
 
 | | Crystal structure of the S228A mutant of the aminopeptidase from Vibrio proteolyticus in complex with leucine | | Descriptor: | Bacterial leucyl aminopeptidase, LEUCINE, SODIUM ION, ... | | Authors: | Ataie, N.J, Hoang, Q.Q, Zahniser, M.P.D, Milne, A, Petsko, G.A, Ringe, D. | | Deposit date: | 2007-10-22 | | Release date: | 2007-11-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Zinc coordination geometry and ligand binding affinity: the structural and kinetic analysis of the second-shell serine 228 residue and the methionine 180 residue of the aminopeptidase from Vibrio proteolyticus.
Biochemistry, 47, 2008
|
|
3B7I
 
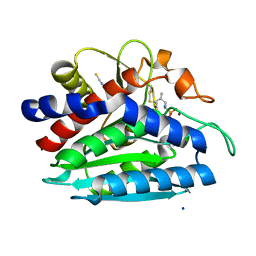 | | Crystal structure of the S228A mutant of the aminopeptidase from Vibrio proteolyticus in complex with leucine phosphonic acid | | Descriptor: | Bacterial leucyl aminopeptidase, LEUCINE, LEUCINE PHOSPHONIC ACID, ... | | Authors: | Ataie, N.J, Hoang, Q.Q, Zahniser, M.P.D, Milne, A, Petsko, G.A, Ringe, D. | | Deposit date: | 2007-10-30 | | Release date: | 2007-11-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Zinc coordination geometry and ligand binding affinity: the structural and kinetic analysis of the second-shell serine 228 residue and the methionine 180 residue of the aminopeptidase from Vibrio proteolyticus.
Biochemistry, 47, 2008
|
|
1IGB
 
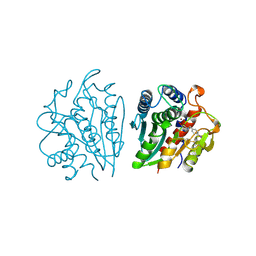 | | AEROMONAS PROTEOLYTICA AMINOPEPTIDASE COMPLEXED WITH THE INHIBITOR PARA-IODO-D-PHENYLALANINE HYDROXAMATE | | Descriptor: | AMINOPEPTIDASE, PARA-IODO-D-PHENYLALANINE HYDROXAMIC ACID, ZINC ION | | Authors: | Chevrier, B, D'Orchymont, H, Schalk, C, Tarnus, C, Moras, D. | | Deposit date: | 1996-02-27 | | Release date: | 1996-08-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of the Aeromonas proteolytica aminopeptidase complexed with a hydroxamate inhibitor. Involvement in catalysis of Glu151 and two zinc ions of the co-catalytic unit.
Eur.J.Biochem., 237, 1996
|
|
2GVL
 
 | | Crystal Structure of Murine NMPRTase | | Descriptor: | Nicotinamide phosphoribosyltransferase | | Authors: | Khan, J.A, Tao, X, Tong, L. | | Deposit date: | 2006-05-02 | | Release date: | 2006-07-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis for the inhibition of human NMPRTase, a novel target for anticancer agents.
Nat.Struct.Mol.Biol., 13, 2006
|
|
1CP6
 
 | | 1-BUTANEBORONIC ACID BINDING TO AEROMONAS PROTEOLYTICA AMINOPEPTIDASE | | Descriptor: | 1-BUTANE BORONIC ACID, PROTEIN (AMINOPEPTIDASE), ZINC ION | | Authors: | Depaola, C.C, Bennett, B, Holz, R.C, Ringe, D, Petsko, G.A. | | Deposit date: | 1999-06-08 | | Release date: | 1999-06-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1-Butaneboronic acid binding to Aeromonas proteolytica aminopeptidase: a case of arrested development.
Biochemistry, 38, 1999
|
|
2GVJ
 
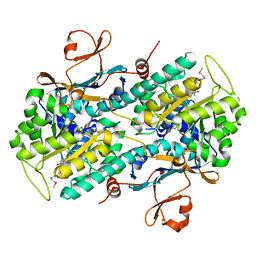 | | Crystal Structure of Human NMPRTase in complex with FK866 | | Descriptor: | (2E)-N-{4-[1-(benzenecarbonyl)piperidin-4-yl]butyl}-3-(pyridin-3-yl)prop-2-enamide, Nicotinamide phosphoribosyltransferase | | Authors: | Khan, J.A, Tao, X, Tong, L. | | Deposit date: | 2006-05-02 | | Release date: | 2006-06-20 | | Last modified: | 2016-11-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis for the inhibition of human NMPRTase, a novel target for anticancer agents.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2H7R
 
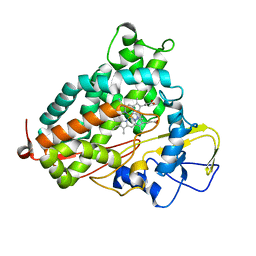 | |
2H7Q
 
 | | Cytochrome P450cam complexed with imidazole | | Descriptor: | Cytochrome P450-cam, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Verras, A, Alian, A, Montellano, P.R. | | Deposit date: | 2006-06-02 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Cytochrome P450 active site plasticity: attenuation of imidazole binding in cytochrome P450cam by an L244A mutation.
Protein Eng.Des.Sel., 19, 2006
|
|
5Y1S
 
 | | Crystal structure of Plasmodium falciparum aminopeptidase N in complex with (S)-2-(3-(3,4-dimethylbenzyl)ureido)-N-hydroxy-4-methylpentanamide | | Descriptor: | (2R)-2-[(3,4-dimethylphenyl)methylcarbamoylamino]-4-methyl-N-oxidanyl-pentanamide, (2S)-2-[(3,4-dimethylphenyl)methylcarbamoylamino]-4-methyl-N-oxidanyl-pentanamide, GLYCEROL, ... | | Authors: | Marapaka, A.K, Zhang, Y, Addlagatta, A. | | Deposit date: | 2017-07-21 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structure of Plasmodium falciparum aminopeptidase N in complex with (S)-2-(3-(3,4-dimethylbenzyl)ureido)-N-hydroxy-4-methylpentanamide
To Be Published
|
|
5YQB
 
 | | Crystal structure of E.coli aminopeptidase N in complex with Puromycin | | Descriptor: | (2R,3R,4S,5S)-4-AMINO-2-[6-(DIMETHYLAMINO)-9H-PURIN-9-YL]-5-(HYDROXYMETHYL)TETRAHYDRO-3-FURANOL, Aminopeptidase N, GLYCEROL, ... | | Authors: | Marapaka, A.K, Ganji, R.J, Reddi, R, Addlagatta, A. | | Deposit date: | 2017-11-06 | | Release date: | 2018-11-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Puromycin, a selective inhibitor of PSA acts as a substrate for other M1 family aminopeptidases: Biochemical and structural basis
Int.J.Biol.Macromol., 165, 2020
|
|
5Y1Q
 
 | | Crystal structure of Plasmodium falciparum aminopeptidase N in complex with (S)-2-(3-(3-chlorobenzyl)ureido)-N-hydroxy-4-methylpentanamide | | Descriptor: | (2S)-2-[(3-chlorophenyl)methylcarbamoylamino]-4-methyl-N-oxidanyl-pentanamide, M1 family aminopeptidase, MAGNESIUM ION, ... | | Authors: | Marapaka, A.K, Zhang, Y, Addlagatta, A. | | Deposit date: | 2017-07-21 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Development of peptidomimetic hydroxamates as PfA-M1 and PfA-M17 dual inhibitors: Biological evaluation and structural characterization by cocrystallization
Chin.Chem.Lett., 33, 2022
|
|
4R5V
 
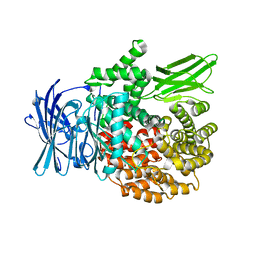 | |
4EYC
 
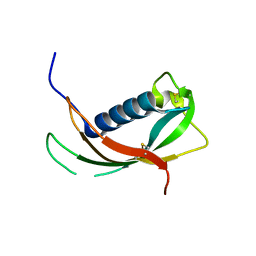 | | Crystal structure of the cathelin-like domain of human cathelicidin LL-37 (hCLD) | | Descriptor: | Cathelicidin antimicrobial peptide | | Authors: | Pazgier, M, Pozharski, E, Toth, E, Lu, W. | | Deposit date: | 2012-05-01 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Analysis of the Pro-Domain of Human Cathelicidin, LL-37.
Biochemistry, 52, 2013
|
|
5YO1
 
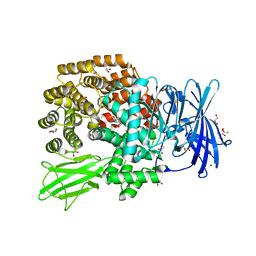 | | Structure of ePepN E298A mutant in complex with Puromycin | | Descriptor: | Aminopeptidase N, GLYCEROL, PUROMYCIN, ... | | Authors: | Ganji, R.J, Reddi, R, Marapaka, A.K, Addlagatta, A. | | Deposit date: | 2017-10-26 | | Release date: | 2018-11-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Puromycin, a selective inhibitor of PSA acts as a substrate for other M1 family aminopeptidases: Biochemical and structural basis
Int.J.Biol.Macromol., 165, 2020
|
|
5Y1V
 
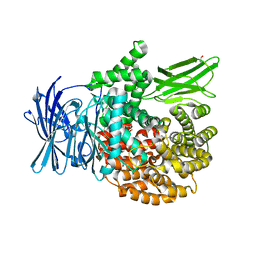 | | Crystal structure of Plasmodium falciparum aminopeptidase N in complex with (S)-2-(3-(2,6-diethylphenyl)ureido)- N-hydroxy-4-methylpentanamide | | Descriptor: | (2R)-2-[(2,6-diethylphenyl)carbamoylamino]-4-methyl-N-oxidanyl-pentanamide, GLYCEROL, M1 family aminopeptidase, ... | | Authors: | Marapaka, A.K, Zhang, Y, Addlagatta, A. | | Deposit date: | 2017-07-21 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structure of Plasmodium falciparum aminopeptidase N in complex with (S)-2-(3-(2,6-diethylphenyl)ureido)- N-hydroxy-4-methylpentanamide
To Be Published
|
|
5Y19
 
 | | Crystal structure of Plasmodium falciparum aminopeptidase N in complex with (S)-2-((2S,3R)-3-amino-2-hydroxy-4-phenylbutanamido)- N-hydroxy-4-methylpentanamide. | | Descriptor: | (2S)-2-[[(2S,3R)-3-azanyl-2-oxidanyl-4-phenyl-butanoyl]amino]-4-methyl-N-oxidanyl-pentanamide, GLYCEROL, M1 family aminopeptidase, ... | | Authors: | Marapaka, A.K, Zhang, Y, Addlagatta, A. | | Deposit date: | 2017-07-19 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of Plasmodium falciparum aminopeptidase N in complex with (S)-2-((2S,3R)-3-amino-2-hydroxy-4-phenylbutanamido)- N-hydroxy-4-methylpentanamide.
To Be Published
|
|
4R8F
 
 | |
5Y1X
 
 | |
5Y1W
 
 | |
4R5X
 
 | |
4ZI6
 
 | |
5YQ1
 
 | | Crystal structure of E.coli aminopeptidase N in complex with O-Methyl-L-tyrosine | | Descriptor: | Aminopeptidase N, GLYCEROL, MALONATE ION, ... | | Authors: | Marapaka, A.K, Ganji, R.J, Reddi, R, Addlagatta, A. | | Deposit date: | 2017-11-04 | | Release date: | 2018-11-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of E.coli aminopeptidase N in complex with O-Methyl-L-tyrosine
To Be Published
|
|
