7DCE
 
 | | Cryo-EM structure of human XKR8-basigin complex bound to Fab fragment | | Descriptor: | 1,2-DILINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Heavy chain of Fab fragment, Isoform 2 of Basigin, ... | | Authors: | Sakuragi, T, Kanai, R, Tsutsumi, A, Narita, H, Onishi, E, Miyazaki, T, Baba, T, Nakagawa, A, Kikkawa, M, Toyoshima, C, Nagata, S. | | Deposit date: | 2020-10-26 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The tertiary structure of the human Xkr8-Basigin complex that scrambles phospholipids at plasma membranes.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7A1H
 
 | | Crystal structure of wild-type CI2 | | Descriptor: | SULFATE ION, Subtilisin-chymotrypsin inhibitor-2A | | Authors: | Olsen, J.G, Teilum, K, Hamborg, L, Roche, J.V. | | Deposit date: | 2020-08-13 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synergistic stabilization of a double mutant in chymotrypsin inhibitor 2 from a library screen in E. coli.
Commun Biol, 4, 2021
|
|
4TKT
 
 | | Streptomyces platensis isomigrastatin ketosynthase domain MgsF KS6 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, AT-less polyketide synthase, CHLORIDE ION, ... | | Authors: | Chang, C, Li, H, Endres, M, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-05-27 | | Release date: | 2014-06-11 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (2.4289 Å) | | Cite: | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6LDH
 
 | |
2XUS
 
 | | Crystal Structure of the BRMS1 N-terminal region | | Descriptor: | BREAST CANCER METASTASIS-SUPPRESSOR 1, CHLORIDE ION, SULFATE ION | | Authors: | Spinola-Amilibia, M, Rivera, J, Ortiz-Lombardia, M, Romero, A, Neira, J.L, Bravo, J. | | Deposit date: | 2010-10-20 | | Release date: | 2011-07-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.912 Å) | | Cite: | The Structure of Brms1 Nuclear Export Signal and Snx6 Interacting Region Reveals a Hexamer Formed by Antiparallel Coiled Coils.
J.Mol.Biol., 411, 2011
|
|
4TZV
 
 | | Co-crystals of the Ternary Complex Containing a T-box Stem I RNA, its Cognate tRNAGly, and B. subtilis YbxF protein, treated by removing lithium sulfate post crystallization | | Descriptor: | Ribosome-associated protein L7Ae-like, T-box stem I, engineered tRNA | | Authors: | Zhang, J, Ferre-D'Amare, A.R. | | Deposit date: | 2014-07-11 | | Release date: | 2014-09-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (5.03 Å) | | Cite: | Dramatic Improvement of Crystals of Large RNAs by Cation Replacement and Dehydration.
Structure, 22, 2014
|
|
7DQE
 
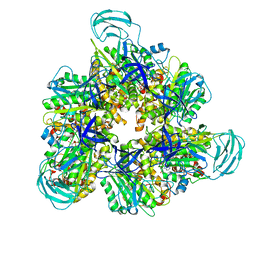 | | Crystal structure of the ADP-bound mutant A(S23C)3B(N64C)3 complex from enterococcus hirae V-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Maruyama, S, Nakamoto, K, Suzuki, K, Mizutani, K, Imai, F.L, Ishizuka-Katsura, Y, Shirouzu, M, Murata, T. | | Deposit date: | 2020-12-23 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.694 Å) | | Cite: | The Combination of High-Speed AFM and X-ray Crystallography Reveals Rotary Catalytic Mechanism of Shaftless V1-ATPase
To Be Published
|
|
7DQD
 
 | | Crystal structure of the AMP-PNP-bound mutant A(S23C)3B(N64C)3 complex from enterococcus hirae V-ATPase | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Maruyama, S, Suzuki, K, Mizutani, K, Imai, F.L, Ishizuka-Katsura, Y, Shirouzu, M, Murata, T. | | Deposit date: | 2020-12-23 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.383 Å) | | Cite: | The combination of high-speed AFM and X-ray crystallography reveals rotary catalysis
To Be Published
|
|
7DQC
 
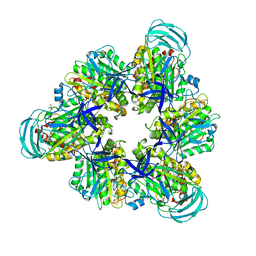 | | Crystal structure of nucleotide-free mutant A(S23C)3B(N64C)3 complex from Enterococcus hirae V-ATPase | | Descriptor: | GLYCEROL, V-type sodium ATPase catalytic subunit A, V-type sodium ATPase subunit B | | Authors: | Maruyama, S, Suzuki, K, Mizutani, K, Imai, F.L, Ishizuka-Katsura, Y, Shirouzu, M, Murata, M. | | Deposit date: | 2020-12-23 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.706 Å) | | Cite: | The combination of high-speed AFM and X-ray crystallography reveals rotary catalytic mechanism of shaftless V1-ATPase
To Be Published
|
|
6V12
 
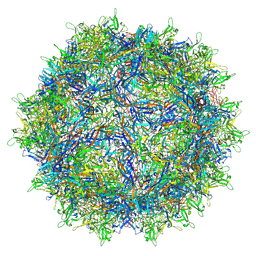 | | Empty AAV8 particles | | Descriptor: | Capsid protein | | Authors: | Mietzsch, M, Agbandje-McKenna, M. | | Deposit date: | 2019-11-19 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Comparative Analysis of the Capsid Structures of AAVrh.10, AAVrh.39, and AAV8.
J.Virol., 94, 2020
|
|
6T66
 
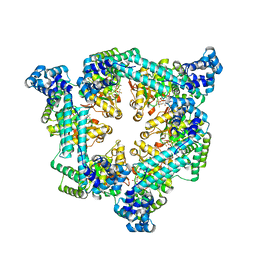 | | Crystal structure of the Vibrio cholerae replicative helicase (DnaB) with GDP-AlF4 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Replicative DNA helicase, ... | | Authors: | Legrand, P, Quevillon-Cheruel, S, Li de la Sierra-Gallay, I, Walbott, H. | | Deposit date: | 2019-10-17 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Study of the DnaB:DciA interplay reveals insights into the primary mode of loading of the bacterial replicative helicase.
Nucleic Acids Res., 49, 2021
|
|
5LMA
 
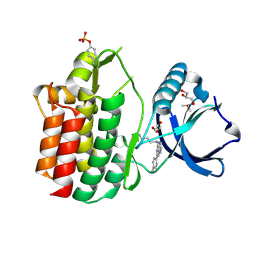 | |
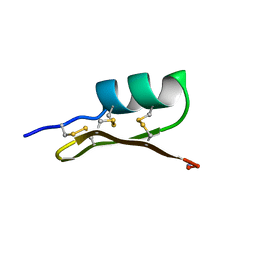
1SCY
 
 | |
6HEQ
 
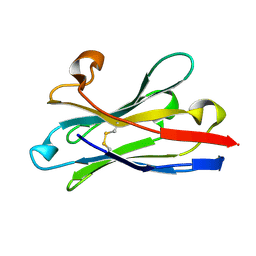 | | Prion nanobody 484 | | Descriptor: | Prion nanobody 484 | | Authors: | Soror, S.H, Abskharon, R.N, Wohlkonig, A. | | Deposit date: | 2018-08-20 | | Release date: | 2019-12-04 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Structural evidence for the critical role of the prion protein hydrophobic region in forming an infectious prion.
Plos Pathog., 15, 2019
|
|
6HF2
 
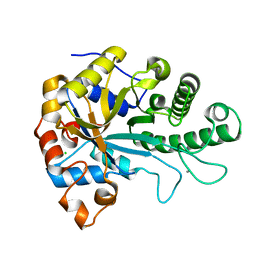 | | The structure of BoMan26B, a GH26 beta-mannanase from Bacteroides ovatus | | Descriptor: | CALCIUM ION, CHLORIDE ION, Glycosyl hydrolase family 26 | | Authors: | Bagenholm, V, Logan, D.T, Stalbrand, H. | | Deposit date: | 2018-08-21 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | A surface-exposed GH26 beta-mannanase fromBacteroides ovatus: Structure, role, and phylogenetic analysis ofBoMan26B.
J.Biol.Chem., 294, 2019
|
|
6HF4
 
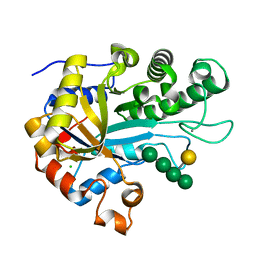 | | The structure of BoMan26B, a GH26 beta-mannanase from Bacteroides ovatus, complexed with G1M4 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Glycosyl hydrolase family 26, ... | | Authors: | Bagenholm, V, Logan, D.T, Stalbrand, H. | | Deposit date: | 2018-08-21 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.781 Å) | | Cite: | A surface-exposed GH26 beta-mannanase fromBacteroides ovatus: Structure, role, and phylogenetic analysis ofBoMan26B.
J.Biol.Chem., 294, 2019
|
|
7WO0
 
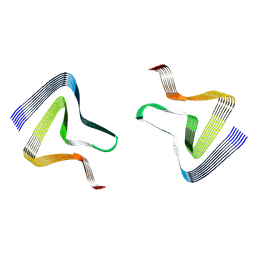 | |
1HI1
 
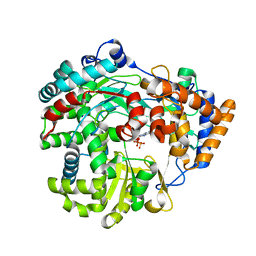 | | RNA dependent RNA polymerase from dsRNA bacteriophage phi6 plus bound NTP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, P2 PROTEIN | | Authors: | Grimes, J.M, Butcher, S.J, Makeyev, E.V, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2001-01-01 | | Release date: | 2001-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A Mechanism for Initiating RNA-Dependent RNA Polymerization
Nature, 410, 2001
|
|
1OAT
 
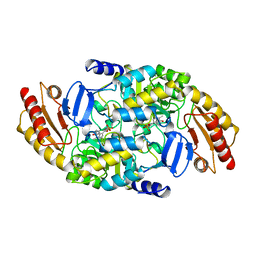 | | ORNITHINE AMINOTRANSFERASE | | Descriptor: | ORNITHINE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Shen, B.W, Schirmer, T, Jansonius, J.N. | | Deposit date: | 1997-03-26 | | Release date: | 1998-04-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human recombinant ornithine aminotransferase.
J.Mol.Biol., 277, 1998
|
|
1HXJ
 
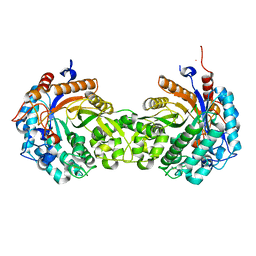 | |
2IS6
 
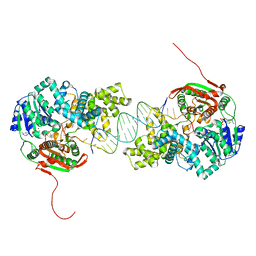 | | Crystal structure of UvrD-DNA-ADPMgF3 ternary complex | | Descriptor: | 5'-D(*CP*GP*AP*GP*CP*AP*CP*TP*GP*CP*AP*GP*TP*GP*CP*TP*CP*GP*TP*TP*GP*TP*TP*AP*T)-3', ADENOSINE-5'-DIPHOSPHATE, DNA helicase II, ... | | Authors: | Yang, W, Lee, J.Y. | | Deposit date: | 2006-10-16 | | Release date: | 2007-01-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke.
Cell(Cambridge,Mass.), 127, 2006
|
|
4UI2
 
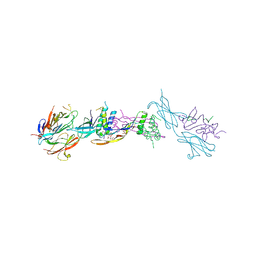 | | Crystal structure of the ternary RGMB-BMP2-NEO1 complex | | Descriptor: | ACETATE ION, BONE MORPHOGENETIC PROTEIN 2, BMP2, ... | | Authors: | Healey, E.G, Bishop, B, Elegheert, J, Bell, C.H, Padilla-Parra, S, Siebold, C. | | Deposit date: | 2015-03-27 | | Release date: | 2015-05-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Repulsive Guidance Molecule is a Structural Bridge between Neogenin and Bone Morphogenetic Protein.
Nat.Struct.Mol.Biol., 22, 2015
|
|
6I3N
 
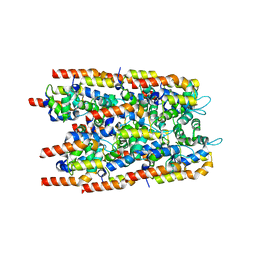 | |
2C9O
 
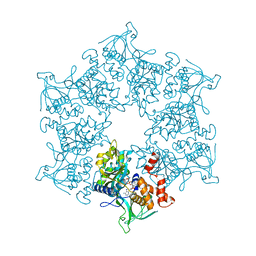 | | 3D Structure of the human RuvB-like helicase RuvBL1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RUVB-LIKE 1 | | Authors: | Matias, P.M, Gorynia, S, Donner, P, Carrondo, M.A. | | Deposit date: | 2005-12-14 | | Release date: | 2006-10-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the human AAA+ protein RuvBL1.
J. Biol. Chem., 281, 2006
|
|
6I1M
 
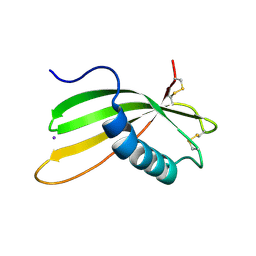 | | Secreted type 1 cystatin from Fasciola hepatica | | Descriptor: | Cystatin, IODIDE ION | | Authors: | Busa, M, Rezacova, P, Pachl, P, Stefanic, S, Mares, M. | | Deposit date: | 2018-10-29 | | Release date: | 2020-08-26 | | Last modified: | 2023-03-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An evolutionary molecular adaptation of an unusual stefin from the liver fluke Fasciola hepatica redefines the cystatin superfamily.
J.Biol.Chem., 299, 2023
|
|
