8JEJ
 
 | | Cryo-EM Structure of Na-dithionite Reduced Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus | | Descriptor: | FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Fructose dehydrogenase cytochrome subunit, ... | | Authors: | Suzuki, Y, Miyata, T, Makino, F, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | Deposit date: | 2023-05-16 | | Release date: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Essential Insight of Direct Electron Transfer-Type Bioelectrocatalysis by Membrane-Bound d-Fructose Dehydrogenase with Structural Bioelectrochemistry.
Acs Catalysis, 13, 2023
|
|
3P1L
 
 | | Crystal structure of Escherichia coli BamB, a lipoprotein component of the beta-barrel assembly machinery complex, native crystals. | | Descriptor: | Lipoprotein yfgL, SODIUM ION | | Authors: | Kim, K.H, Paetzel, M. | | Deposit date: | 2010-09-30 | | Release date: | 2010-12-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Escherichia coli BamB, a lipoprotein component of the beta-barrel assembly machinery complex, native crystals
J.Mol.Biol., 406, 2011
|
|
4B52
 
 | | Crystal structure of Gentlyase, the neutral metalloprotease of Paenibacillus polymyxa | | Descriptor: | BACILLOLYSIN, CALCIUM ION, N-ALPHA-L-RHAMNOPYRANOSYLOXY(HYDROXYPHOSPHINYL)-L-LEUCYL-L-TRYPTOPHAN, ... | | Authors: | Ruf, A, Stihle, M, Benz, J, Schmidt, M, Sobek, H. | | Deposit date: | 2012-08-02 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structure of Gentlyase, the Neutral Metalloprotease of Paenibacillus Polymyxa
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3P7Z
 
 | |
8IQ8
 
 | | Crystal structure of 3,4-dihydroxyphenylacetate 2,3-dioxygenase (DHPAO) from Acinetobacter baumannii | | Descriptor: | 3,4-dihydroxyphenylacetate 2,3-dioxygenase, SODIUM ION | | Authors: | Chitnumsub, P, Maenpuen, S. | | Deposit date: | 2023-03-16 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and biochemical characterization of an extradiol 3,4-dihydroxyphenylacetate 2,3-dioxygenase from Acinetobacter baumannii.
Arch.Biochem.Biophys., 747, 2023
|
|
3P9O
 
 | | Aerobic ternary complex of urate oxidase with azide and chloride | | Descriptor: | AZIDE ION, CHLORIDE ION, SODIUM ION, ... | | Authors: | Gabison, L, Colloc'H, N, El Hajji, M, Castro, B, Chiadmi, M, Prange, T. | | Deposit date: | 2010-10-18 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Azide and Cyanide Show Different Inhibition Modes to Urate Oxidase
To be Published
|
|
7JXN
 
 | |
4B2J
 
 | | COMPLEXES OF DODECIN WITH FLAVIN AND FLAVIN-LIKE LIGANDS | | Descriptor: | CHLORIDE ION, DODECIN, MAGNESIUM ION, ... | | Authors: | Staudt, H, Hoesl, M, Dreuw, A, Serdjukow, S, Oesterhelt, D, Budisa, N, Wachtveitl, J, Grininger, M. | | Deposit date: | 2012-07-16 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Directed Manipulation of a Flavoprotein Photocycle.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
8J2L
 
 | | Crystal structure of a bright green fluorescent protein (StayGold) with double mutations (N137A, Y187F) in jellyfish Cytaeis uchidae from Biortus | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, SODIUM ION, ... | | Authors: | Wu, J, Wang, F, Gui, W, Cheng, W, Yang, Y. | | Deposit date: | 2023-04-14 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a bright green fluorescent protein (StayGold) in jellyfish Cytaeis uchidae from Biortus
To Be Published
|
|
8J2H
 
 | | Crystal structure of a bright green fluorescent protein (StayGold) with single mutation (N137A) in jellyfish Cytaeis uchidae from Biortus | | Descriptor: | GLYCEROL, SODIUM ION, StayGold(N137A) | | Authors: | Wu, J, Wang, F, Gui, W, Cheng, W, Yang, Y. | | Deposit date: | 2023-04-14 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a bright green fluorescent protein (StayGold) in jellyfish Cytaeis uchidae from Biortus
To Be Published
|
|
3QVX
 
 | | L-myo-inositol 1-phosphate synthase from Archaeoglobus fulgidus mutant K367A | | Descriptor: | GLYCEROL, Myo-inositol-1-phosphate synthase (Ino1), NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Neelon, K, Roberts, M.F, Stec, B. | | Deposit date: | 2011-02-26 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a trapped catalytic intermediate suggests that forced atomic proximity drives the catalysis of mIPS.
Biophys.J., 101, 2011
|
|
3QNG
 
 | |
3QR7
 
 | | Crystal structure of the C-terminal fragment of the bacteriophage P2 membrane-piercing protein gpV | | Descriptor: | Baseplate assembly protein V, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Browning, C, Shneider, M, Leiman, P.G. | | Deposit date: | 2011-02-17 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Phage pierces the host cell membrane with the iron-loaded spike.
Structure, 20, 2012
|
|
3PLG
 
 | | urate oxidase under 1.0 MPa / 10 bars pressure of equimolar mixture xenon : nitrous oxide | | Descriptor: | 8-AZAXANTHINE, NITROUS OXIDE, SODIUM ION, ... | | Authors: | Marassio, G, Colloc'h, N, Prange, T, Abraini, J.H. | | Deposit date: | 2010-11-15 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Pressure-response analysis of anesthetic gases xenon and nitrous oxide on urate oxidase: a crystallographic study.
Faseb J., 25, 2011
|
|
3PG0
 
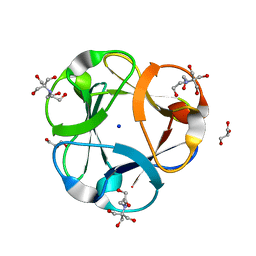 | | Crystal structure of designed 3-fold symmetric protein, ThreeFoil | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, SODIUM ION, ... | | Authors: | Lobsanov, Y.D, Broom, A, Howell, P.L, Rose, D.R, Meiering, E.M. | | Deposit date: | 2010-10-29 | | Release date: | 2011-12-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Modular evolution and the origins of symmetry: reconstruction of a three-fold symmetric globular protein.
Structure, 20, 2012
|
|
7K16
 
 | | Tamana Bat Virus xrRNA1 | | Descriptor: | MAGNESIUM ION, RNA (51-MER), SODIUM ION | | Authors: | Jones, R.A, Kieft, J.S. | | Deposit date: | 2020-09-07 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Different tertiary interactions create the same important 3D features in a distinct flavivirus xrRNA.
Rna, 27, 2021
|
|
3ZN6
 
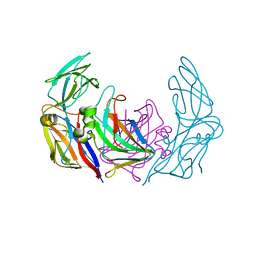 | | VP16-VP17 complex, a complex of the two major capsid proteins of bacteriophage P23-77 | | Descriptor: | CHLORIDE ION, SODIUM ION, VP16, ... | | Authors: | Rissanen, I, Grimes, J.M, Pawlowski, A, Mantynen, S, Harlos, K, Bamford, J.K.H, Stuart, D.I. | | Deposit date: | 2013-02-13 | | Release date: | 2013-05-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Bacteriophage P23-77 Capsid Protein Structures Reveal the Archetype of an Ancient Branch from a Major Virus Lineage.
Structure, 21, 2013
|
|
7JZ0
 
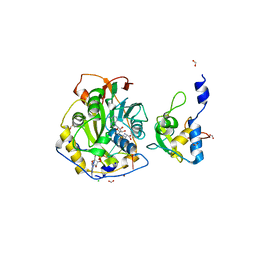 | | Crystal Structure of SARS-CoV-2 Nsp16/10 Heterodimer in Complex with (m7GpppA2m)pUpUpApApA (Cap-1) and S-Adenosyl-L-homocysteine (SAH). | | Descriptor: | 2'-O-methyltransferase, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Kiryukhina, O, Brunzelle, J.S, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-09-01 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of SARS-CoV-2 2'-O-methyltransferase heterodimer with RNA Cap analog and sulfates bound reveals new strategies for structure-based inhibitor design
Biorxiv, 2020
|
|
7K96
 
 | |
7KDS
 
 | |
8ITZ
 
 | | Crystal structure of human Galectin-3 in complex with small molecule inhibitor | | Descriptor: | 2-[(2R,3R,4S,5R,6R)-2-(3,4-dichlorophenyl)sulfanyl-6-(hydroxymethyl)-5-oxidanyl-4-[4-[3,4,5-tris(fluoranyl)phenyl]-1,2,3-triazol-1-yl]oxan-3-yl]oxyethanoic acid, CHLORIDE ION, Galectin-3, ... | | Authors: | Jinal, S, Amit, K, Ghosh, K. | | Deposit date: | 2023-03-23 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Discovery and Exploration of Monosaccharide Linked Dimers to Target Fibrosis
To Be Published
|
|
7KBU
 
 | | Structure of Hevin FS-EC | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Machius, M, Fan, S, Rudenko, G. | | Deposit date: | 2020-10-03 | | Release date: | 2021-02-10 | | Last modified: | 2021-07-14 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Interplay between hevin, SPARC, and MDGAs: Modulators of neurexin-neuroligin transsynaptic bridges.
Structure, 29, 2021
|
|
7K97
 
 | | Human DNA polymerase beta dGDP product complex with Mn2+ | | Descriptor: | DNA (5'-D(*CP*CP*GP*AP*CP*CP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*CP*G)-3'), DNA (5'-D(P*GP*TP*CP*GP*G)-3'), ... | | Authors: | Varela, F.A, Freudenthal, B.D. | | Deposit date: | 2020-09-28 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanism of Deoxyguanosine Diphosphate Insertion by Human DNA Polymerase beta.
Biochemistry, 60, 2021
|
|
7K15
 
 | | Crystal structure of the Human Leukotriene B4 Receptor 1 in Complex with Selective Antagonist MK-D-046 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, FLAVIN MONONUCLEOTIDE, HEXAETHYLENE GLYCOL, ... | | Authors: | Michaelian, N, Han, G.W, Cherezov, V. | | Deposit date: | 2020-09-07 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structural insights on ligand recognition at the human leukotriene B4 receptor 1.
Nat Commun, 12, 2021
|
|
8IZJ
 
 | |
