7OZG
 
 | |
6QUS
 
 | | HsCKK (human CAMSAP1) decorated 13pf taxol-GDP microtubule | | Descriptor: | Calmodulin-regulated spectrin-associated protein 1, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Atherton, J.M, Luo, Y, Xiang, S, Yang, C, Jiang, K, Stangier, M, Vemu, A, Cook, A, Wang, S, Roll-Mecak, A, Steinmetz, M.O, Akhmanova, A, Baldus, M, Moores, C.A. | | Deposit date: | 2019-02-28 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural determinants of microtubule minus end preference in CAMSAP CKK domains.
Nat Commun, 10, 2019
|
|
3KU7
 
 | | Crystal structure of Helicobacter pylori MinE, a cell division topological specificity factor | | Descriptor: | Cell division topological specificity factor | | Authors: | Kang, G.B, Song, H.E, Kim, M.K, Eom, S.H. | | Deposit date: | 2009-11-26 | | Release date: | 2010-05-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of Helicobacter pylori MinE, a cell division topological specificity factor
Mol.Microbiol., 76, 2010
|
|
3TOI
 
 | |
3U1C
 
 | |
6DJY
 
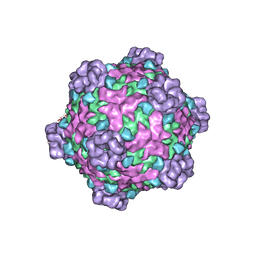 | | Fako virus | | Descriptor: | Clamp protein, Major capsid protein, Turret protein | | Authors: | Kaelber, J.T, Jiang, W, Weaver, S.C, Auguste, A.J, Chiu, W. | | Deposit date: | 2018-05-27 | | Release date: | 2019-06-05 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | The polymerase organization of Fako virus
To be Published
|
|
7NPQ
 
 | |
8SLO
 
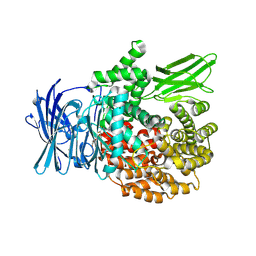 | | Plasmodium falciparum M1 aminopeptidase bound to selective inhibitor MIPS2673 | | Descriptor: | 2-hydroxy-N-[(1R)-2-(hydroxyamino)-2-oxo-1-(3',4',5'-trifluoro[1,1'-biphenyl]-4-yl)ethyl]-2-methylpropanamide, GLYCEROL, M1 family aminopeptidase, ... | | Authors: | McGowan, S.M, Drinkwater, N. | | Deposit date: | 2023-04-24 | | Release date: | 2024-04-03 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Chemoproteomics validates selective targeting of Plasmodium M1 alanyl aminopeptidase as an antimalarial strategy.
Elife, 13, 2024
|
|
8SVL
 
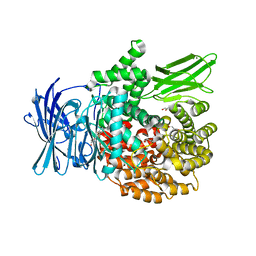 | |
8SVM
 
 | | Plasmodium falciparum M17 aminopeptidase bound to MMV1557817 | | Descriptor: | CARBONATE ION, Leucine aminopeptidase, N-[(1R)-2-(hydroxyamino)-2-oxo-1-(3',4',5'-trifluoro[1,1'-biphenyl]-4-yl)ethyl]-3,3-dimethylbutanamide, ... | | Authors: | McGowan, S, Drinkwater, N. | | Deposit date: | 2023-05-17 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterisation of a novel antimalarial agent targeting haemaglobin digestion that shows cross-species reactivity and excellent in vivo properties.
Mbio, 2024
|
|
4BH9
 
 | |
1CP6
 
 | | 1-BUTANEBORONIC ACID BINDING TO AEROMONAS PROTEOLYTICA AMINOPEPTIDASE | | Descriptor: | 1-BUTANE BORONIC ACID, PROTEIN (AMINOPEPTIDASE), ZINC ION | | Authors: | Depaola, C.C, Bennett, B, Holz, R.C, Ringe, D, Petsko, G.A. | | Deposit date: | 1999-06-08 | | Release date: | 1999-06-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1-Butaneboronic acid binding to Aeromonas proteolytica aminopeptidase: a case of arrested development.
Biochemistry, 38, 1999
|
|
5W2D
 
 | |
5W30
 
 | | Crystal structure of mutant CJ YCEI protein (CJ-N48C) with monobromobimane guest structure | | Descriptor: | 3-(bromomethyl)-2,5,6-trimethyl-1H,7H-pyrazolo[1,2-a]pyrazole-1,7-dione, Putative periplasmic protein, SULFATE ION, ... | | Authors: | Huber, T.R, Snow, C.D. | | Deposit date: | 2017-06-07 | | Release date: | 2018-01-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Installing Guest Molecules at Specific Sites within Scaffold Protein Crystals.
Bioconjug. Chem., 29, 2018
|
|
5W39
 
 | | Crystal structure of mutant CJ YCEI protein (CJ-N182C) with monobromobimane guest structure | | Descriptor: | 3-(bromomethyl)-2,5,6-trimethyl-1H,7H-pyrazolo[1,2-a]pyrazole-1,7-dione, EICOSANE, Polyisoprenoid-binding protein, ... | | Authors: | Huber, T.R, Snow, C.D. | | Deposit date: | 2017-06-07 | | Release date: | 2018-01-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Installing Guest Molecules at Specific Sites within Scaffold Protein Crystals.
Bioconjug. Chem., 29, 2018
|
|
5W3A
 
 | |
5W32
 
 | |
5W3C
 
 | |
5VTS
 
 | | Dehaloperoxidase B Y28F mutant | | Descriptor: | Dehaloperoxidase B, GLYCEROL, OXYGEN MOLECULE, ... | | Authors: | Carey, L.M, Ghiladi, R.A. | | Deposit date: | 2017-05-18 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.571 Å) | | Cite: | Probing the Structure-Function Relationship of a Multifunctional Enzyme using Crystallographic Diffraction Methods
Thesis, North Carolina State University, 2017
|
|
5W2Z
 
 | |
5W2K
 
 | |
5W2R
 
 | |
5W2X
 
 | |
5W31
 
 | |
5W3B
 
 | |
