5WN5
 
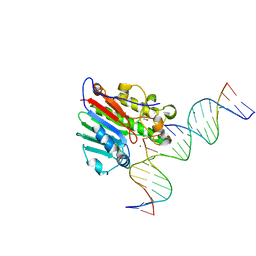 | | APE1 exonuclease substrate complex with a C/T mismatch and Mn2+ | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*(DV3))-3'), DNA (5'-D(*GP*GP*AP*TP*CP*CP*GP*TP*CP*GP*AP*TP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Freudenthal, B.D, Whitaker, A.M. | | Deposit date: | 2017-07-31 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular snapshots of APE1 proofreading mismatches and removing DNA damage.
Nat Commun, 9, 2018
|
|
5F8Z
 
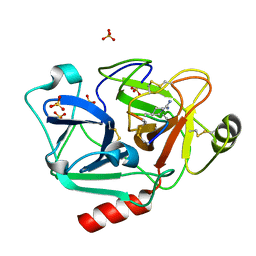 | | The crystal structure of human Plasma Kallikrein in complex with its peptide inhibitor pkalin-1 | | Descriptor: | CYS-PRO-ALA-ARG-PHE-M70-ALA-LEU-PHE-CYS, Plasma kallikrein LIGHT CHAIN, SULFATE ION, ... | | Authors: | Xu, M, Jiang, L, Xu, P, Luo, Z, Andreasen, P, Huang, M. | | Deposit date: | 2015-12-09 | | Release date: | 2016-12-14 | | Last modified: | 2025-09-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of human Plasma Kallikrein in complex with its peptide inhibitor pkalin-1
To Be Published
|
|
4UZN
 
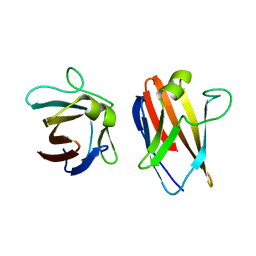 | | The native structure of the family 46 carbohydrate-binding module (CBM46) of endo-beta-1,4-glucanase B (Cel5B) from Bacillus halodurans | | Descriptor: | ENDO-BETA-1,4-GLUCANASE (CELULASE B) | | Authors: | Venditto, I, Santos, H, Ferreira, L.M.A, Sakka, K, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2014-09-05 | | Release date: | 2015-02-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Family 46 Carbohydrate-Binding Modules Contribute to the Enzymatic Hydrolysis of Xyloglucan and Beta-1,3-1,4-Glucans Through Distinct Mechanisms.
J.Biol.Chem., 290, 2015
|
|
4W9P
 
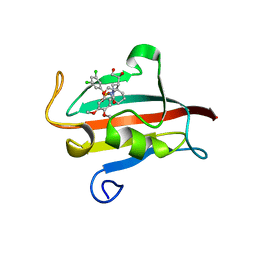 | | The Fk1 domain of FKBP51 in complex with (1S,5S,6R)-10-[(3,5-dichlorophenyl)sulfonyl]-5-[(1S)-1,2-dihydroxyethyl]-3-[2-(3,4-dimethoxyphenoxy)ethyl]-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1S,5S,6R)-10-[(3,5-dichlorophenyl)sulfonyl]-5-[(1S)-1,2-dihydroxyethyl]-3-[2-(3,4-dimethoxyphenoxy)ethyl]-3,10-diazabicyclo[4.3.1]decan-2-one, ACETATE ION, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Pomplun, S, Wang, Y, Kirschner, K, Kozany, C, Bracher, A, Hausch, F. | | Deposit date: | 2014-08-27 | | Release date: | 2014-12-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Rational Design and Asymmetric Synthesis of Potent and Neurotrophic Ligands for FK506-Binding Proteins (FKBPs).
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4URK
 
 | | PI3Kg in complex with AZD6482 | | Descriptor: | 2-[[(1R)-1-(7-methyl-2-morpholin-4-yl-4-oxidanylidene-pyrido[1,2-a]pyrimidin-9-yl)ethyl]amino]benzoic acid, PHOSPHATIDYLINOSITOL 4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT GAMMA ISOFORM | | Authors: | Giordanetto, F, Barlaam, B, Berglund, S, Edman, K, Karlsson, O, Lindberg, J, Nylander, S, Inghardt, T. | | Deposit date: | 2014-06-30 | | Release date: | 2014-10-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of 9-(1-Phenoxyethyl)-2-Morpholino-4-Oxo-Pyrido[1, 2-A]Pyrimidine-7-Carboxamides as Oral Pi3Kbeta Inhibitors, Useful as Antiplatelet Agents.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
5LUC
 
 | | Crystal structure of the D183N variant of human Alanine:Glyoxylate Aminotransferase major allele (AGT-Ma) at 1.8 Angstrom; internal aldimine with PLP in the active site | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PYRIDOXAL-5'-PHOSPHATE, Serine--pyruvate aminotransferase | | Authors: | Giardina, G, Cutruzzola, F, Cellini, B, Borri Voltattorni, C, Montioli, R. | | Deposit date: | 2016-09-08 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Radiation damage at the active site of human alanine:glyoxylate aminotransferase reveals that the cofactor position is finely tuned during catalysis.
Sci Rep, 7, 2017
|
|
5M3A
 
 | | Crystal structure of BRD4 BROMODOMAIN 1 IN COMPLEX WITH LIGAND 2 | | Descriptor: | 1,2-ETHANEDIOL, 3-methyl-6-(1-methyl-5-phenoxy-pyrazol-4-yl)-[1,2,4]triazolo[4,3-b]pyridazine, Bromodomain-containing protein 4 | | Authors: | Kessler, D, Mayer, M, Engelhardt, H, Wolkerstorfer, B, Geist, L. | | Deposit date: | 2016-10-14 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Direct NMR Probing of Hydration Shells of Protein Ligand Interfaces and Its Application to Drug Design.
J. Med. Chem., 60, 2017
|
|
5M1R
 
 | | X-ray structure of human G166D PGK-1 mutant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Phosphoglycerate kinase 1 | | Authors: | Ilari, A, Cipollone, A, Fiorillo, A, Petrosino, M. | | Deposit date: | 2016-10-10 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | The phosphoglycerate kinase 1 variants found in carcinoma cells display different catalytic activity and conformational stability compared to the native enzyme.
PLoS ONE, 13, 2018
|
|
8C7Y
 
 | | Crystal structure of BRAF V600E in complex with a hybrid compound 6 | | Descriptor: | 1,2-ETHANEDIOL, NITRATE ION, Serine/threonine-protein kinase B-raf, ... | | Authors: | Chaikuad, A, Bonnet, P, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-01-17 | | Release date: | 2023-02-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Design, synthesis and characterisation of a novel type II B-RAF paradox breaker inhibitor.
Eur.J.Med.Chem., 250, 2023
|
|
8C7X
 
 | | Crystal structure of BRAF in complex with a hybrid compound 6 | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, Serine/threonine-protein kinase B-raf, ... | | Authors: | Chaikuad, A, Bonnet, P, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-01-17 | | Release date: | 2023-02-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Design, synthesis and characterisation of a novel type II B-RAF paradox breaker inhibitor.
Eur.J.Med.Chem., 250, 2023
|
|
5LVS
 
 | | Self-assembled protein-aromatic foldamer complexes with 2:3 and 2:2:1 stoichiometries | | Descriptor: | Aromatic foldamer, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Jewginski, M, LANGLOIS D'ESTAINTOT, B, Granier, T, Huc, Y. | | Deposit date: | 2016-09-14 | | Release date: | 2017-03-08 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Self-Assembled Protein-Aromatic Foldamer Complexes with 2:3 and 2:2:1 Stoichiometries.
J. Am. Chem. Soc., 139, 2017
|
|
5WT9
 
 | | Complex structure of PD-1 and nivolumab-Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of Nivolumab, Light Chain of Nivolumab, ... | | Authors: | Tan, S, Zhang, H, Chai, Y, Song, H, Tong, Z, Wang, Q, Qi, J, Wong, G, Zhu, X, Liu, W.J, Gao, S, Wang, Z, Shi, Y, Yang, F, Gao, G.F, Yan, J. | | Deposit date: | 2016-12-10 | | Release date: | 2017-02-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | An unexpected N-terminal loop in PD-1 dominates binding by nivolumab.
Nat Commun, 8, 2017
|
|
5MD9
 
 | | The structure of the mature HIV-1 CA hexameric lattice with curvature parameters: tilt=17, twist=6 | | Descriptor: | Capsid protein p24 C-terminal domain, Capsid protein p24 N-terminal domain | | Authors: | Mattei, S, Glass, B, Hagen, W.J.H, Kraeusslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2016-11-10 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | The structure and flexibility of conical HIV-1 capsids determined within intact virions.
Science, 354, 2016
|
|
5MD7
 
 | | The structure of the mature HIV-1 CA hexameric lattice with curvature parameters: tilt=11, twist=-12 | | Descriptor: | Capsid protein p24 | | Authors: | Mattei, S, Glass, B, Hagen, W.J.H, Kraeusslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2016-11-10 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | The structure and flexibility of conical HIV-1 capsids determined within intact virions.
Science, 354, 2016
|
|
5MD3
 
 | | The structure of the mature HIV-1 CA hexameric lattice with curvature parameters: tilt=11, twist=12 | | Descriptor: | Capsid protein p24 | | Authors: | Mattei, S, Glass, B, Hagen, W.J.H, Kraeusslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2016-11-10 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | The structure and flexibility of conical HIV-1 capsids determined within intact virions.
Science, 354, 2016
|
|
5MDE
 
 | | The structure of the mature HIV-1 CA hexameric lattice with curvature parameters: tilt=23, twist=0 | | Descriptor: | Capsid protein p24 C-terminal domain, Capsid protein p24 N-terminal domain | | Authors: | Mattei, S, Glass, B, Hagen, W.J.H, Kraeusslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2016-11-10 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | The structure and flexibility of conical HIV-1 capsids determined within intact virions.
Science, 354, 2016
|
|
5MDA
 
 | | The structure of the mature HIV-1 CA hexameric lattice with curvature parameters: tilt=17, twist=0 | | Descriptor: | Gag protein | | Authors: | Mattei, S, Glass, B, Hagen, W.J.H, Kraeusslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2016-11-10 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | The structure and flexibility of conical HIV-1 capsids determined within intact virions.
Science, 354, 2016
|
|
5M6Z
 
 | | The X-ray structure of human M189I PGK-1 mutant in partially closed conformation | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ilari, A, Fiorillo, A, Petrosino, M, Cipollone, A. | | Deposit date: | 2016-10-26 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The phosphoglycerate kinase 1 variants found in carcinoma cells display different catalytic activity and conformational stability compared to the native enzyme.
PLoS ONE, 13, 2018
|
|
5MD8
 
 | | The structure of the mature HIV-1 CA hexameric lattice with curvature parameters: tilt=17, twist=12 | | Descriptor: | Gag protein | | Authors: | Mattei, S, Glass, B, Hagen, W.J.H, Kraeusslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2016-11-10 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.6 Å) | | Cite: | The structure and flexibility of conical HIV-1 capsids determined within intact virions.
Science, 354, 2016
|
|
4UZ6
 
 | | STRUCTURE OF THE WNT DEACYLASE NOTUM - CRYSTAL FORM V - SOS COMPLEX - 1.9A | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Zebisch, M, Jones, E.Y. | | Deposit date: | 2014-09-04 | | Release date: | 2015-02-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Notum Deacylates Wnt Proteins to Suppress Signalling Activity.
Nature, 519, 2015
|
|
5MD4
 
 | | The structure of the mature HIV-1 CA hexameric lattice with curvature parameters: tilt=11, twist=6 | | Descriptor: | Capsid protein p24 | | Authors: | Mattei, S, Glass, B, Hagen, W.J.H, Kraeusslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2016-11-10 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | The structure and flexibility of conical HIV-1 capsids determined within intact virions.
Science, 354, 2016
|
|
5MDG
 
 | | The structure of the mature HIV-1 CA hexameric lattice with curvature parameters: tilt=29, twist=0 | | Descriptor: | Gag protein | | Authors: | Mattei, S, Glass, B, Hagen, W.J.H, Kraeusslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2016-11-10 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.7 Å) | | Cite: | The structure and flexibility of conical HIV-1 capsids determined within intact virions.
Science, 354, 2016
|
|
5G1O
 
 | | Aspartate transcarbamoylase domain of human CAD in apo form | | Descriptor: | 1,2-ETHANEDIOL, CAD protein, GLYCEROL | | Authors: | Ruiz-Ramos, A, Grande-Garcia, A, Moreno-Morcillo, M.D, Ramon-Maiques, S. | | Deposit date: | 2016-03-29 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and Functional Characterization of Human Aspartate Transcarbamoylase, the Target of the Anti-Tumoral Drug Pala.
Structure, 24, 2016
|
|
7LT9
 
 | |
7LT8
 
 | | Crystal structure of Ras suppressor-1 | | Descriptor: | Ras suppressor protein 1 | | Authors: | Fukuda, K, Qin, J. | | Deposit date: | 2021-02-19 | | Release date: | 2021-04-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.76000249 Å) | | Cite: | Molecular basis for Ras suppressor-1 binding to PINCH-1 in focal adhesion assembly.
J.Biol.Chem., 296, 2021
|
|
