1ABB
 
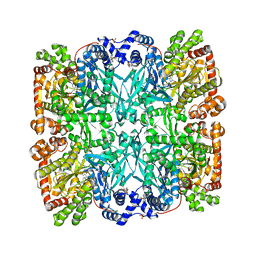 | | CONTROL OF PHOSPHORYLASE B CONFORMATION BY A MODIFIED COFACTOR: CRYSTALLOGRAPHIC STUDIES ON R-STATE GLYCOGEN PHOSPHORYLASE RECONSTITUTED WITH PYRIDOXAL 5'-DIPHOSPHATE | | Descriptor: | GLYCOGEN PHOSPHORYLASE B, INOSINIC ACID, PYRIDOXAL-5'-DIPHOSPHATE, ... | | Authors: | Leonidas, D.D, Oikonomakos, N.G, Papageorgiou, A.C, Acharya, K.R, Barford, D, Johnson, L.N. | | Deposit date: | 1992-04-09 | | Release date: | 1993-10-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Control of phosphorylase b conformation by a modified cofactor: crystallographic studies on R-state glycogen phosphorylase reconstituted with pyridoxal 5'-diphosphate.
Protein Sci., 1, 1992
|
|
3H1J
 
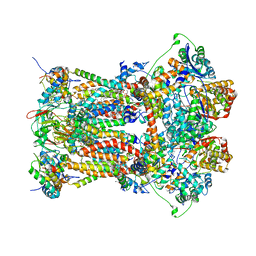 | | Stigmatellin-bound cytochrome bc1 complex from chicken | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CARDIOLIPIN, Coenzyme Q10, ... | | Authors: | Zhang, Z, Huang, L, Shulmeister, V.M, Chi, Y.-I, Kim, K.K, Hung, L.-W, Crofts, A.R, Berry, E.A, Kim, S.-H. | | Deposit date: | 2009-04-12 | | Release date: | 2009-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Electron Transfer by Domain Movement in Cytochrome Bc1
Nature, 392, 1998
|
|
3L73
 
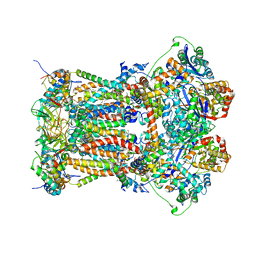 | | Cytochrome BC1 complex from chicken with triazolone inhibitor | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 4-[7-(3,3-dimethylbut-1-yn-1-yl)naphthalen-1-yl]-5-methoxy-2-methyl-2,4-dihydro-3H-1,2,4-triazol-3-one, CARDIOLIPIN, ... | | Authors: | Huang, L, Berry, E.A. | | Deposit date: | 2009-12-27 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Famoxadone and related inhibitors bind like methoxy acrylate inhibitors in the Qo site of the BC1 compl and fix the rieske iron-sulfur protein in a positio close to but distinct from that seen with stigmatellin and other "DISTAL" Qo inhibitors.
To be Published
|
|
2E4F
 
 | |
4UZU
 
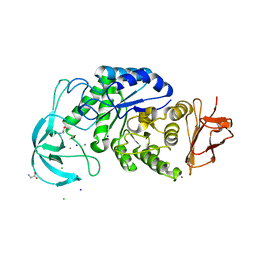 | | Three-dimensional structure of a variant `Termamyl-like' Geobacillus stearothermophilus alpha-amylase at 1.9 A resolution | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Offen, W.A, Anderson, C, Borchert, T.V, Wilson, K.S, Davies, G.J. | | Deposit date: | 2014-09-09 | | Release date: | 2015-01-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Three-Dimensional Structure of a Variant `Termamyl-Like' Geobacillus Stearothermophilus Alpha-Amylase at 1.9 A Resolution
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4UTF
 
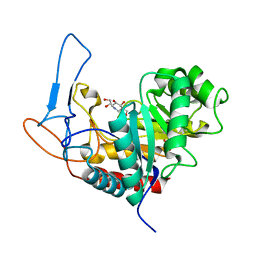 | | Structure of the GH99 endo-alpha-mannosidase from Bacteroides xylanisolvens in complex with mannose-alpha-1,3-isofagomine and alpha- 1,2-mannobiose | | Descriptor: | 1,2-ETHANEDIOL, 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, GLYCOSYL HYDROLASE FAMILY 71, ... | | Authors: | Cuskin, F, Lowe, E.C, Temple, M.J, Zhu, Y, Pudlo, N.A, Cameron, E.A, Urs, K, Thompson, A.J, Cartmell, A, Rogowski, A, Tolbert, T, Piens, K, Bracke, D, Vervecken, W, Hakki, Z, Speciale, G, Munoz-Munoz, J.L, Pena, M.J, McLean, R, Suits, M.D, Boraston, A.B, Atherly, T, Ziemer, C.J, Williams, S.J, Davies, G.J, Abbott, D.W, Martens, E.C, Gilbert, H.J. | | Deposit date: | 2014-07-21 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Human Gut Bacteroidetes Can Utilize Yeast Mannan Through a Selfish Mechanism.
Nature, 517, 2015
|
|
8VQU
 
 | | Crystal Structure of Dehaloperoxidase B in complex with substrate 1,4-cyclohexadiene | | Descriptor: | Dehaloperoxidase B, GLYCEROL, OXYGEN MOLECULE, ... | | Authors: | de Seerano, V.S, Yun, D, Ghiladi, R.A. | | Deposit date: | 2024-01-19 | | Release date: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Crystal Structure of Dehaloperoxidase B in complex with substrate 1,4-cyclohexadiene
To Be Published
|
|
1DJ6
 
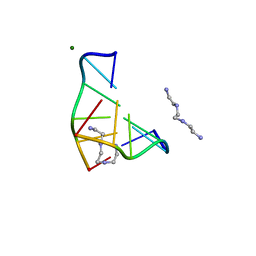 | | COMPLEX OF A Z-DNA HEXAMER, D(CG)3, WITH SYNTHETIC POLYAMINE AT ROOM TEMPERATURE | | Descriptor: | 5'-D(*CP*GP*CP*GP*CP*G)-3', MAGNESIUM ION, N,N'-BIS(2-AMINOETHYL)-1,2-ETHANEDIAMINE | | Authors: | Ohishi, H, Tomita, K.-i, Nakanishi, I, Ohtsuchi, M, Hakoshima, T, Rich, A. | | Deposit date: | 1999-12-01 | | Release date: | 1999-12-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | The crystal structure of N1-[2-(2-amino-ethylamino)-ethyl]-ethane-1,2-diamine (polyamines) binding to the minor groove of d(CGCGCG)2, hexamer at room temperature
FEBS Lett., 523, 2002
|
|
8VOU
 
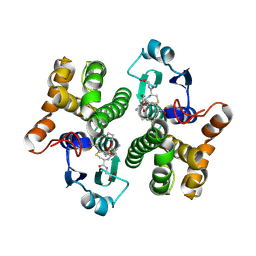 | | Human glutathione transferase M1-1 in complex with the adduct between glutathione and nitrooleic acid | | Descriptor: | GLUTATHIONE, Glutathione S-transferase Mu 1, L-gamma-glutamyl-S-[(8R,9S)-1-carboxy-9-nitroheptadecan-8-yl]-L-cysteinylglycine | | Authors: | Larrieux, N, Steglich, M, Dalla Rizza, J, Buschiazzo, A, Turell, L. | | Deposit date: | 2024-01-16 | | Release date: | 2025-03-12 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Human glutathione transferases catalyze the reaction between glutathione and nitrooleic acid.
J.Biol.Chem., 301, 2025
|
|
8VWE
 
 | | HIV-1 neutralizing antibody D5_AR bound to HIV-1 gp41 coiled-coil pocket IQN17 | | Descriptor: | Neutralizing antibody D5_AR Heavy Chain, Neutralizing antibody D5_AR Light Chain, Transmembrane protein gp41 IQN17 peptide | | Authors: | Filsinger Interrante, M.V, Tang, S, Fernandez, D, Kim, P.S. | | Deposit date: | 2024-02-01 | | Release date: | 2025-07-02 | | Last modified: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Utilizing Machine Learning to Improve Neutralization Potency of an HIV-1 Antibody Targeting the gp41 N-Heptad Repeat.
Acs Chem.Biol., 20, 2025
|
|
8VQG
 
 | | X-ray crystal structure of Can f 1 | | Descriptor: | MALONATE ION, Major allergen Can f 1 | | Authors: | Khatri, K, Geoffrey, M.A, Pedersen, L.C, Champan, M.D, Pomes, A, Chruszcz, M. | | Deposit date: | 2024-01-18 | | Release date: | 2025-01-22 | | Last modified: | 2025-09-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Human IgE monoclonal antibodies define two unusual epitopes trapping dog allergen Can f 1 in different conformations.
Protein Sci., 34, 2025
|
|
8VQF
 
 | | X-ray crystal structure of natural Can f 1 in complex with human IgE 1J11 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IgE 1J11 Heavy chain, IgE 1J11 Light chain, ... | | Authors: | Khatri, K, Ball, A, Richardson, C.M, Smith, S.A, Champan, M.D, Pomes, A, Chruszcz, M. | | Deposit date: | 2024-01-18 | | Release date: | 2025-01-22 | | Last modified: | 2025-09-03 | | Method: | X-RAY DIFFRACTION (3.109 Å) | | Cite: | Human IgE monoclonal antibodies define two unusual epitopes trapping dog allergen Can f 1 in different conformations.
Protein Sci., 34, 2025
|
|
1DFS
 
 | | SOLUTION STRUCTURE OF THE ALPHA-DOMAIN OF MOUSE METALLOTHIONEIN-1 | | Descriptor: | CADMIUM ION, METALLOTHIONEIN-1 | | Authors: | Zangger, K, Oz, G, Otvos, J.D, Armitage, I.M. | | Deposit date: | 1999-11-20 | | Release date: | 1999-12-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of mouse [Cd7]-metallothionein-1 by homonuclear and heteronuclear NMR spectroscopy.
Protein Sci., 8, 1999
|
|
1DCY
 
 | |
8VRP
 
 | | HIV-CA Disulfide linked Hexamer bound to 4-Quinazolinone Scaffold inhibitor | | Descriptor: | IODIDE ION, N-[(1S)-1-[(3P,7M)-3-{4-chloro-3-[(ethanesulfonyl)amino]-1-(2,2,2-trifluoroethyl)-1H-indazol-7-yl}-7-(3-fluoro-4-formylphenyl)-4-oxo-3,4-dihydroquinazolin-2-yl]-2-(3,5-difluorophenyl)ethyl]-2-[3-(trifluoromethyl)-5,6-dihydrocyclopenta[c]pyrazol-1(4H)-yl]acetamide, Spacer peptide 1 | | Authors: | Goldstone, D.C, Walsham, L. | | Deposit date: | 2024-01-22 | | Release date: | 2025-07-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of 4-Quinazolinone-bearing Phenylalanine Derivatives as HIV-1 Capsid Inhibitors with Potent and Insensitivity Towards Clinically Relevant Resistant Mutations
To Be Published
|
|
8W37
 
 | |
1DFT
 
 | | SOLUTION STRUCTURE OF THE BETA-DOMAIN OF MOUSE METALLOTHIONEIN-1 | | Descriptor: | CADMIUM ION, METALLOTHIONEIN-1 | | Authors: | Zangger, K, Oz, G, Otvos, J.D, Armitage, I.M. | | Deposit date: | 1999-11-20 | | Release date: | 1999-12-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of mouse [Cd7]-metallothionein-1 by homonuclear and heteronuclear NMR spectroscopy.
Protein Sci., 8, 1999
|
|
7M67
 
 | |
1DMS
 
 | | STRUCTURE OF DMSO REDUCTASE | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, DMSO REDUCTASE, MOLYBDENUM (IV)OXIDE | | Authors: | Schneider, F, Loewe, J, Huber, R, Schindelin, H, Kisker, C, Knaeblein, J. | | Deposit date: | 1996-09-03 | | Release date: | 1998-07-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure of dimethyl sulfoxide reductase from Rhodobacter capsulatus at 1.88 A resolution.
J.Mol.Biol., 263, 1996
|
|
1DG3
 
 | | STRUCTURE OF HUMAN GUANYLATE BINDING PROTEIN-1 IN NUCLEOTIDE FREE FORM | | Descriptor: | PROTEIN (INTERFERON-INDUCED GUANYLATE-BINDING PROTEIN 1) | | Authors: | Prakash, B, Praefcke, G.J.K, Renault, L, Wittinghofer, A, Herrmann, C. | | Deposit date: | 1999-11-23 | | Release date: | 2000-10-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of human guanylate-binding protein 1 representing a unique class of GTP-binding proteins.
Nature, 403, 2000
|
|
1DB2
 
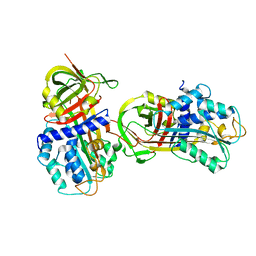 | | CRYSTAL STRUCTURE OF NATIVE PLASMINOGEN ACTIVATOR INHIBITOR-1 | | Descriptor: | PLASMINOGEN ACTIVATOR INHIBITOR-1 | | Authors: | Nar, H, Bauer, M, Stassen, J.M, Lang, D, Gils, A, Declerck, P. | | Deposit date: | 1999-11-02 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Plasminogen activator inhibitor 1. Structure of the native serpin, comparison to its other conformers and implications for serpin inactivation.
J.Mol.Biol., 297, 2000
|
|
1DBF
 
 | |
1DDK
 
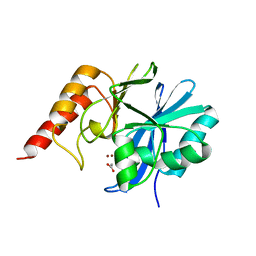 | | CRYSTAL STRUCTURE OF IMP-1 METALLO BETA-LACTAMASE FROM PSEUDOMONAS AERUGINOSA | | Descriptor: | ACETIC ACID, IMP-1 METALLO BETA-LACTAMASE, ZINC ION | | Authors: | Concha, N.O, Janson, C.A, Rowling, P, Pearson, S, Cheever, C.A, Clarke, B.P, Lewis, C, Galleni, M, Frere, J.M, Payne, D.J, Bateson, J.H, Abdel-Meguid, S.S. | | Deposit date: | 1999-11-10 | | Release date: | 2000-11-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of the IMP-1 metallo beta-lactamase from Pseudomonas aeruginosa and its complex with a mercaptocarboxylate inhibitor: binding determinants of a potent, broad-spectrum inhibitor.
Biochemistry, 39, 2000
|
|
1DGI
 
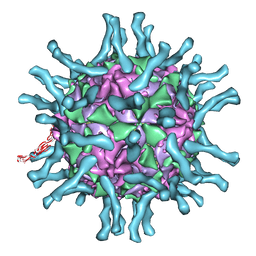 | | Cryo-EM structure of human poliovirus(serotype 1)complexed with three domain CD155 | | Descriptor: | POLIOVIRUS RECEPTOR, VP1, VP2, ... | | Authors: | He, Y, Bowman, V.D, Mueller, S, Bator, C.M, Bella, J, Peng, X, Baker, T.S, Wimmer, E, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 1999-11-24 | | Release date: | 2000-01-24 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (22 Å) | | Cite: | Interaction of the poliovirus receptor with poliovirus.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1DJW
 
 | | PHOSPHOINOSITIDE-SPECIFIC PHOSPHOLIPASE C-DELTA1 FROM RAT COMPLEXED WITH INOSITOL-2-METHYLENE-1,2-CYCLIC-MONOPHOSPHONATE | | Descriptor: | ACETATE ION, CALCIUM ION, INOSITOL-2-METHYLENE-1,2-CYCLIC-MONOPHOSPHATE, ... | | Authors: | Essen, L.-O, Perisic, O, Williams, R.L. | | Deposit date: | 1996-08-24 | | Release date: | 1997-07-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural mapping of the catalytic mechanism for a mammalian phosphoinositide-specific phospholipase C.
Biochemistry, 36, 1997
|
|
