8XM5
 
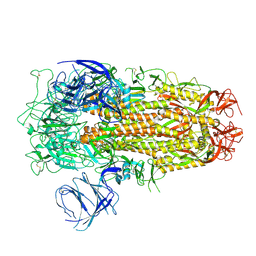 | | Cryo-EM structure of SARS-CoV-2 Omicron EG.5 spike protein(6P), RBD-closed state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Li, L.J, Gu, Y.H, Shi, K.Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2023-12-27 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
8XNF
 
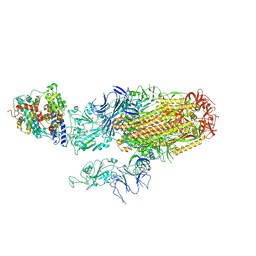 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2.86 spike protein(6P) in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Li, L.J, Gu, Y.H, Shi, K.Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2023-12-29 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
8XN2
 
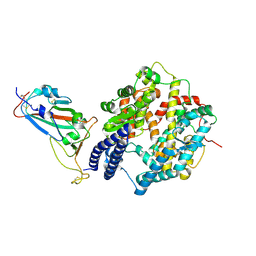 | | SARS-CoV-2 Omicron EG.5.1 RBD in complex with human ACE2 (local refined from the spike protein) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Li, L.J, Gu, Y.H, Shi, K.Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2023-12-28 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
8Y18
 
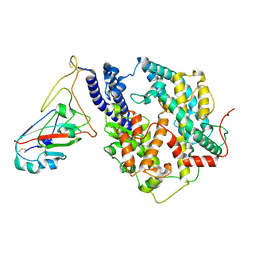 | | Cryo-EM structure of SARS-CoV-2 Omicron JN.1 RBD in complex with human ACE2 (local refinement from the spike protein) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Li, L.J, Gu, Y.H, Qi, J.X, Gao, G.F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-07-03 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
8Y6A
 
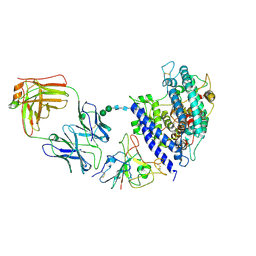 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2.86 RBD in complex with human ACE2 and S309 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Li, L.J, Gu, Y.H, Qi, J.X, Gao, G.F. | | Deposit date: | 2024-02-02 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
8Y81
 
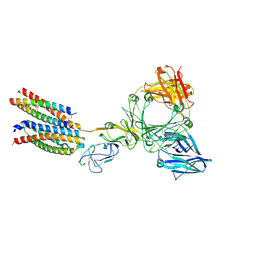 | | Structure of the ige-fc bound to its high affinity receptor fc(epsilon)ri | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Du, S, Deng, M.J, Xiao, J.Y. | | Deposit date: | 2024-02-05 | | Release date: | 2024-07-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structural insights into the high-affinity IgE receptor Fc epsilon RI complex.
Nature, 633, 2024
|
|
8YM1
 
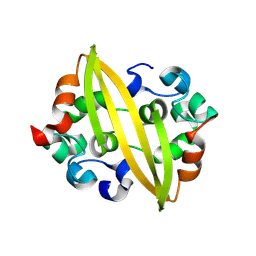 | | Structure of SADS-CoV Virus Nucleocapsid Protein | | Descriptor: | nucleocapsid phosphoprotein | | Authors: | Zhang, Y, Wu, F, Xu, W. | | Deposit date: | 2024-03-08 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unraveling the assembly mechanism of SADS-CoV virus nucleocapsid protein: insights from RNA binding, dimerization, and epitope diversity profiling.
J.Virol., 98, 2024
|
|
8Y3Q
 
 | | ASFV p72 in complex with Fab F11 | | Descriptor: | B646L, Heavy chain of F11, Light chain of F11 | | Authors: | Wang, X, Fu, W, Yu, Q. | | Deposit date: | 2024-01-29 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | p72 antigenic mapping reveals a potential supersite of vulnerability for African swine fever virus.
Cell Discov, 10, 2024
|
|
8Y3O
 
 | | ASFV p72 in complex with Fab B1 | | Descriptor: | B646L, Heavy chain of B1, Light chain of B1 | | Authors: | Wang, X, Fu, W, Yu, Q. | | Deposit date: | 2024-01-29 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | p72 antigenic mapping reveals a potential supersite of vulnerability for African swine fever virus.
Cell Discov, 10, 2024
|
|
8Y3R
 
 | | ASFV p72 in complex with Fab H3 | | Descriptor: | B646L, Heavy chain of H3, Light chain of H3 | | Authors: | Wang, X, Fu, W, Yu, Q. | | Deposit date: | 2024-01-29 | | Release date: | 2024-09-04 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | p72 antigenic mapping reveals a potential supersite of vulnerability for African swine fever virus.
Cell Discov, 10, 2024
|
|
8Y3P
 
 | | ASFV p72 in complex with Fab C9 | | Descriptor: | B646L, Heavy chain of C9, Light chain of C9 | | Authors: | Wang, X, Fu, W, Yu, Q. | | Deposit date: | 2024-01-29 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | p72 antigenic mapping reveals a potential supersite of vulnerability for African swine fever virus.
Cell Discov, 10, 2024
|
|
8Y3N
 
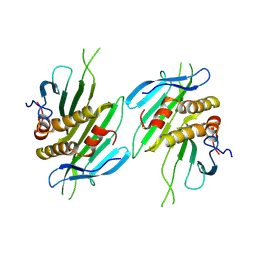 | | The self-assembled nanotube of CPC46A/Q70C | | Descriptor: | Capsid protein | | Authors: | Yang, M, Rao, G, Li, L, Qi, L, Ma, C, Zhang, H, Gong, J, Wei, B, Zhang, X.E, Chen, G, Cao, S, Li, F. | | Deposit date: | 2024-01-29 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Transformation of a Viral Capsid from Nanocages to Nanotubes and Then to Hydrogels: Redirected Self-Assembly and Effects on Immunogenicity.
Acs Nano, 18, 2024
|
|
8Y3T
 
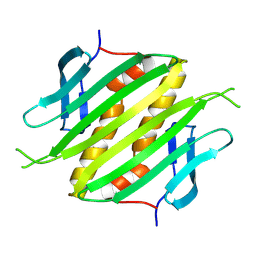 | | The self-assembled nanotube of CPC46A/Q70H | | Descriptor: | Capsid protein | | Authors: | Yang, M, Rao, G, Li, L, Qi, L, Ma, C, Zhang, H, Gong, J, Wei, B, Zhang, X.E, Chen, G, Cao, S, Li, F. | | Deposit date: | 2024-01-29 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Transformation of a Viral Capsid from Nanocages to Nanotubes and Then to Hydrogels: Redirected Self-Assembly and Effects on Immunogenicity.
Acs Nano, 18, 2024
|
|
8Y3V
 
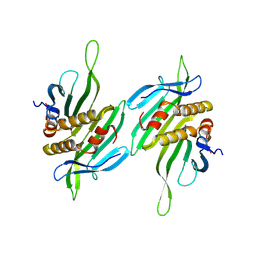 | | The self-assembled nanotube of CPC46A/Q70V | | Descriptor: | Capsid protein | | Authors: | Yang, M, Rao, G, Li, L, Qi, L, Ma, C, Zhang, H, Gong, J, Wei, B, Zhang, X.E, Chen, G, Cao, S, Li, F. | | Deposit date: | 2024-01-29 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Transformation of a Viral Capsid from Nanocages to Nanotubes and Then to Hydrogels: Redirected Self-Assembly and Effects on Immunogenicity.
Acs Nano, 18, 2024
|
|
8XK2
 
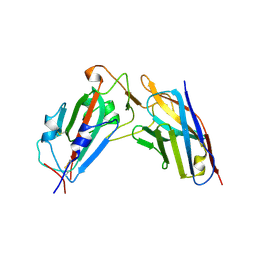 | | A neutralizing nanobody VHH60 against wt SARS-CoV-2 | | Descriptor: | Spike protein S1, VHH60 nanobody | | Authors: | Lu, Y, Guo, H, Ji, X, Yang, H. | | Deposit date: | 2023-12-22 | | Release date: | 2024-07-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | A broad neutralizing nanobody against SARS-CoV-2 engineered from an approved drug.
Cell Death Dis, 15, 2024
|
|
8XKI
 
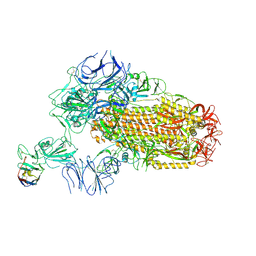 | | A neutralizing nanobody VHH60 against wt SARS-CoV-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y, Guo, H, Ji, X, Yang, H. | | Deposit date: | 2023-12-23 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A broad neutralizing nanobody against SARS-CoV-2 engineered from an approved drug.
Cell Death Dis, 15, 2024
|
|
8ZGU
 
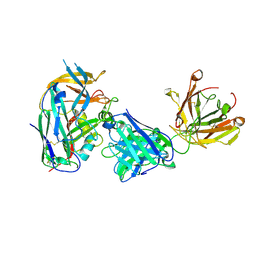 | |
9BJM
 
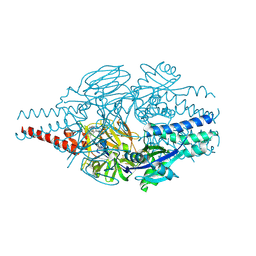 | | Crystal Structure of Inhibitor 5c in Complex with Prefusion RSV F Glycoprotein | | Descriptor: | 1'-{[5-chloro-1-(4,4,4-trifluorobutyl)-1H-1,3-benzimidazol-2-yl]methyl}-1-(methanesulfonyl)spiro[azetidine-3,3'-indol]-2'(1'H)-one, Prefusion RSV F (DS-CAV1),Envelope glycoprotein | | Authors: | Shaffer, P.L, Milligan, C, Abeywickrema, P. | | Deposit date: | 2024-04-25 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Spiro-Azetidine Oxindoles as Long-Acting Injectables for Pre-Exposure Prophylaxis against Respiratory Syncytial Virus Infections.
J.Med.Chem., 67, 2024
|
|
8XLV
 
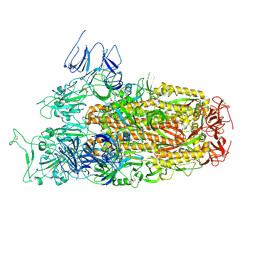 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2.86 spike protein(6P), 1-RBD-up state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Li, L.J, Gu, Y.H, Shi, K.Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2023-12-26 | | Release date: | 2024-07-03 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
8Y5J
 
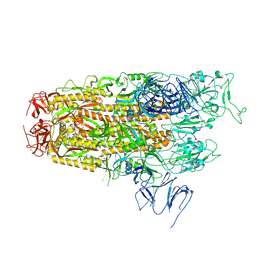 | | Cryo-EM structure of SARS-CoV-2 Omicron JN.1 spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Li, L.J, Gu, Y.H, Qi, J.X, Gao, G.F. | | Deposit date: | 2024-01-31 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
8Y16
 
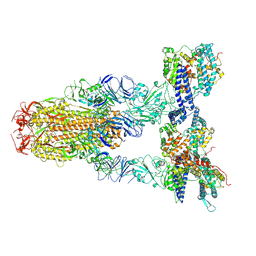 | | Cryo-EM structure of SARS-CoV-2 Omicron JN.1 spike protein in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Li, L.J, Gu, Y.H, Qi, J.X, Gao, G.F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
8XJ0
 
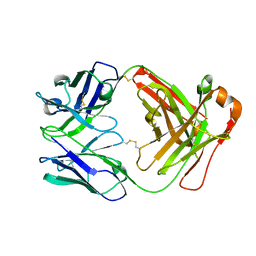 | | Crystal structure of AmFab mutant - P40C/E165C (Light chain), G10C/P210C(Heavy chain) | | Descriptor: | Adalimumab Fab Heavy chain, Adalimumab Fab Light chain | | Authors: | Senda, M, Yoshikawa, M, Nakamura, H, Ohkuri, T, Senda, T. | | Deposit date: | 2023-12-20 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Stabilization of adalimumab Fab through the introduction of disulfide bonds between the variable and constant domains.
Biochem.Biophys.Res.Commun., 700, 2024
|
|
8ZL9
 
 | | ASFV p72 in complex with Fab G6 | | Descriptor: | B646L, G6 Heavy chain, G6 Light chain | | Authors: | Wang, X, Fu, W, Yu, Q. | | Deposit date: | 2024-05-17 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.36 Å) | | Cite: | p72 antigenic mapping reveals a potential supersite of vulnerability for African swine fever virus.
Cell Discov, 10, 2024
|
|
7L7K
 
 | |
7L7F
 
 | |
