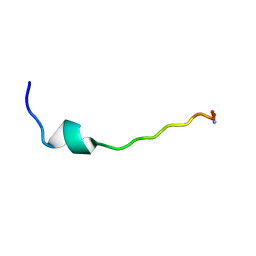
4U1C
 
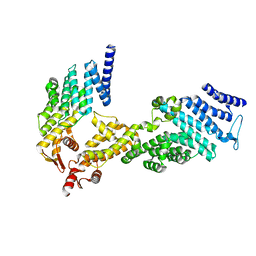 | |
1XUE
 
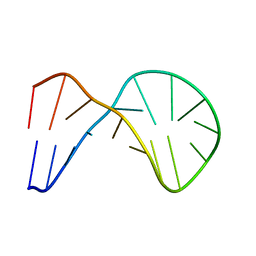 | |
2HTG
 
 | | Structural and functional characterization of TM VII of the NHE1 isoform of the Na+/H+ exchanger | | Descriptor: | NHE1 isoform of Na+/H+ exchanger 1 | | Authors: | Rainey, J.K, Ding, J, Xu, C, Fliegel, L, Sykes, B.D. | | Deposit date: | 2006-07-25 | | Release date: | 2006-08-08 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structural and functional characterization of transmembrane segment VII of the Na(+)/H(+) exchanger isoform 1
J.Biol.Chem., 281, 2006
|
|
5Y7F
 
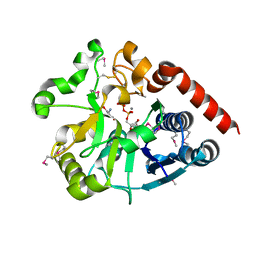 | | Crystal structure of catalytic domain of UGGT (UDP-bound form) from Thermomyces dupontii | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, UGGT, ... | | Authors: | Satoh, T, Song, C, Zhu, T, Toshimori, T, Murata, K, Hayashi, Y, Kamikubo, H, Uchihashi, T, Kato, K. | | Deposit date: | 2017-08-17 | | Release date: | 2017-09-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Visualisation of a flexible modular structure of the ER folding-sensor enzyme UGGT.
Sci Rep, 7, 2017
|
|
1LRY
 
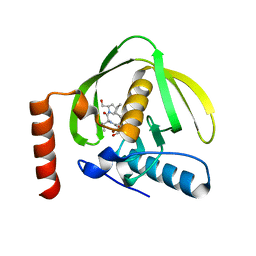 | | Crystal Structure of P. aeruginosa Peptide Deformylase Complexed with Antibiotic Actinonin | | Descriptor: | ACTINONIN, PEPTIDE deformylase, ZINC ION | | Authors: | Guilloteau, J.-P, Mathieu, M, Giglione, C, Blanc, V, Dupuy, A, Chevrier, M, Gil, P, Famechon, A, Meinnel, T, Mikol, V. | | Deposit date: | 2002-05-16 | | Release date: | 2002-07-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structures of four peptide deformylases bound to the antibiotic actinonin reveal two distinct types: a platform for the structure-based design of antibacterial agents.
J.Mol.Biol., 320, 2002
|
|
4M24
 
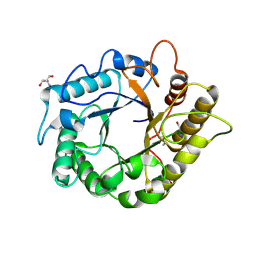 | | Crystal structure of the endo-1,4-glucanase, RBcel1, in complex with cellobiose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endoglucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Delsaute, M, Berlemont, R, Van elder, D, Galleni, M, Bauvois, C. | | Deposit date: | 2013-08-05 | | Release date: | 2014-12-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.386 Å) | | Cite: | Characterisation of two GH family 5 cellulases required for bacterial cellulose production
To be Published
|
|
7S3B
 
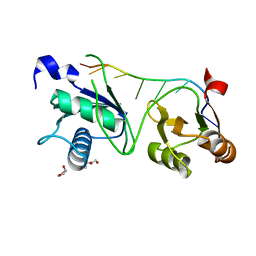 | |
7S3C
 
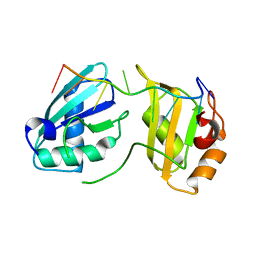 | |
5Y4F
 
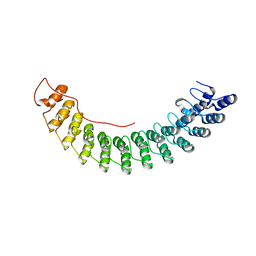 | | Crystal Structure of AnkB Ankyrin Repeats R13-24 in complex with autoinhibition segment AI-c | | Descriptor: | ACETATE ION, Ankyrin-2, CALCIUM ION | | Authors: | Chen, K, Li, J, Wang, C, Wei, Z, Zhang, M. | | Deposit date: | 2017-08-03 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.953 Å) | | Cite: | Autoinhibition of ankyrin-B/G membrane target bindings by intrinsically disordered segments from the tail regions.
Elife, 6, 2017
|
|
7S3A
 
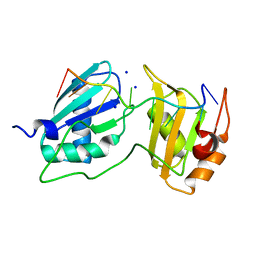 | |
4MH2
 
 | | Crystal structure of Bovine Mitochondrial Peroxiredoxin III | | Descriptor: | CITRIC ACID, Thioredoxin-dependent peroxide reductase, mitochondrial | | Authors: | Cao, Z, McGow, D.P, Shepherd, C, Lindsay, J.G. | | Deposit date: | 2013-08-29 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Improved Catenated Structures of Bovine Peroxiredoxin III F190L Reveal Details of Ring-Ring Interactions and a Novel Conformational State.
Plos One, 10, 2015
|
|
6OZZ
 
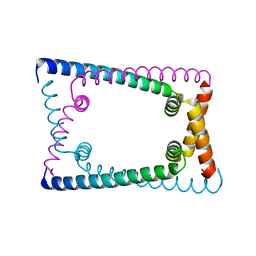 | | N terminally deleted GapR crystal structure from C. crescentus | | Descriptor: | UPF0335 protein CC_3319 | | Authors: | Tarry, M, Harmel, C, Taylor, J.A, Marczynski, G.T, Schmeing, T.M. | | Deposit date: | 2019-05-16 | | Release date: | 2019-11-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.304 Å) | | Cite: | Structures of GapR reveal a central channel which could accommodate B-DNA.
Sci Rep, 9, 2019
|
|
1MJD
 
 | | Structure of N-terminal domain of human doublecortin | | Descriptor: | DOUBLECORTIN | | Authors: | Kim, M.H, Cierpicki, T, Derewenda, U, Krowarsch, D, Feng, Y, Devedjiev, Y, Dauter, Z, Walsh, C.A, Otlewski, J, Bushweller, J.H, Derewenda, Z.S. | | Deposit date: | 2002-08-27 | | Release date: | 2003-04-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The DCX-domain Tandems of Doublecortin and Doublecortin-like Kinase
Nat.Struct.Biol., 10, 2003
|
|
5E2J
 
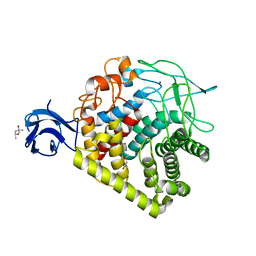 | | Crystal structure of single mutant thermostable endoglucanase (D468A) from Alicyclobacillus acidocaldarius | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Endoglucanase, ... | | Authors: | Hsiao, Y.Y, Wang, H.J, Tseng, C.P. | | Deposit date: | 2015-10-01 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Polarity Alteration of a Calcium Site Induces a Hydrophobic Interaction Network and Enhances Cel9A Endoglucanase Thermostability.
Appl.Environ.Microbiol., 82, 2016
|
|
4X2Y
 
 | | Crystal structure of a chimeric Murine Norovirus NS6 protease (inactive C139A mutant) in which the P4-P4 prime residues of the cleavage junction in the extended C-terminus have been replaced by the corresponding residues from the NS2-3 junction. | | Descriptor: | NS6 Protease,NS6 Protease | | Authors: | Leen, E.N, Pfeil, M.-P, Fernandes, H, Curry, S. | | Deposit date: | 2014-11-27 | | Release date: | 2015-02-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.417 Å) | | Cite: | Structure determination of Murine Norovirus NS6 proteases with C-terminal extensions designed to probe protease-substrate interactions.
Peerj, 3, 2015
|
|
8UQ7
 
 | | Alzheimer's disease PHF complexed with PET ligand MK-6240 | | Descriptor: | 6-fluoro-3-(1H-pyrrolo[2,3-c]pyridin-1-yl)isoquinolin-5-amine, Microtubule-associated protein tau | | Authors: | Kunach, P, Shahmoradian, S.H, Diamond, M.I, Rosa-Neto, P. | | Deposit date: | 2023-10-23 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.31 Å) | | Cite: | Cryo-EM structure of Alzheimer's disease tau filaments with PET ligand MK-6240.
Nat Commun, 15, 2024
|
|
4X6G
 
 | |
6Z2B
 
 | |
5E8G
 
 | |
5GVA
 
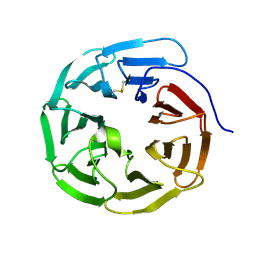 | | WD40 domain of human AND-1 | | Descriptor: | WD repeat and HMG-box DNA-binding protein 1 | | Authors: | Guan, C.C, Li, J. | | Deposit date: | 2016-09-04 | | Release date: | 2017-04-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | The structure and polymerase-recognition mechanism of the crucial adaptor protein AND-1 in the human replisome
J. Biol. Chem., 292, 2017
|
|
1LWA
 
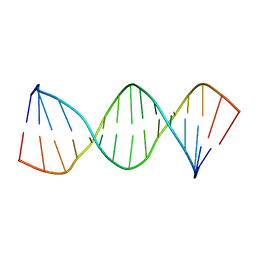 | | Solution Structure of SRY_DNA | | Descriptor: | 5'-D(*CP*TP*GP*AP*AP*CP*AP*AP*TP*CP*AP*CP*CP*CP*C)-3', 5'-D(*GP*GP*GP*GP*TP*GP*AP*TP*TP*GP*TP*TP*CP*AP*G)-3' | | Authors: | Masse, J.E, Wong, B, Yen, Y.-M, Allain, F.H.-T, Johnson, R.C, Feigon, J. | | Deposit date: | 2002-05-30 | | Release date: | 2002-10-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The S. cerevisiae architectural HMGB protein NHP6A complexed with DNA: DNA and protein conformational changes upon binding
J.Mol.Biol., 323, 2002
|
|
4WWB
 
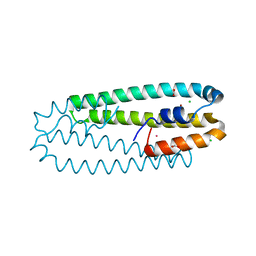 | | High-resolution structure of the Ni-bound form of the Y135F mutant of C. metallidurans CnrXs | | Descriptor: | CARBON DIOXIDE, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Volbeda, A, Coves, J, Maillard, A.P, Kuennemann, S, Grosse, C, Schleuder, G, Petit-Haertlein, I, de Rosny, E, Nies, D.H. | | Deposit date: | 2014-11-10 | | Release date: | 2015-02-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Response of CnrX from Cupriavidus metallidurans CH34 to nickel binding.
Metallomics, 7, 2015
|
|
4X2X
 
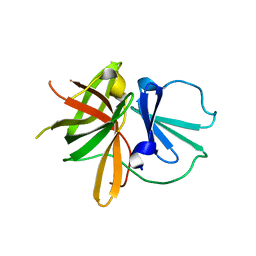 | |
1M62
 
 | | Solution structure of the BAG domain from BAG4/SODD | | Descriptor: | BAG-family molecular chaperone regulator-4 | | Authors: | Briknarova, K, Takayama, S, Homma, S, Baker, K, Cabezas, E, Hoyt, D.W, Li, Z, Satterthwait, A.C, Ely, K.R. | | Deposit date: | 2002-07-11 | | Release date: | 2002-07-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | BAG4/SODD protein contains a short BAG domain.
J.Biol.Chem., 277, 2002
|
|
6WCU
 
 | | Crystal structure of coiled coil region of human septin 5 | | Descriptor: | Septin-5 | | Authors: | Cabrejos, D.A.L, Cavini, I, Sala, F.A, Valadares, N.F, Pereira, H.M, Brandao-Neto, J, Nascimento, A.F.Z, Uson, I, Araujo, A.P.U, Garratt, R.C. | | Deposit date: | 2020-03-31 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Orientational Ambiguity in Septin Coiled Coils and its Structural Basis.
J.Mol.Biol., 433, 2021
|
|
