8H4Z
 
 | |
8H3U
 
 | | Inhibitor-bound EP, polyA model | | Descriptor: | Enteropeptidase catalytic light chain, Enteropeptidase non-catalytic heavy chain | | Authors: | Ding, Z.Y, Huang, H.J. | | Deposit date: | 2022-10-09 | | Release date: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Cryo-EM structures reveal the activation and substrate recognition mechanism of human enteropeptidase.
Nat Commun, 13, 2022
|
|
6W3Y
 
 | | Crystal structure of ligand-binding domain of Campylobacter jejuni chemoreceptor Tlp3 in complex with L-alanine | | Descriptor: | ALANINE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Khan, M.F, Machuca, M.A, Rahman, M.M, Roujeinikova, A. | | Deposit date: | 2020-03-09 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structure-Activity Relationship Study Reveals the Molecular Basis for Specific Sensing of Hydrophobic Amino Acids by theCampylobacter jejuniChemoreceptor Tlp3.
Biomolecules, 10, 2020
|
|
8H3S
 
 | | Substrate-bound EP, polyA model | | Descriptor: | Enteropeptidase catalytic light chain, Enteropeptidase non-catalytic heavy chain, Serine protease 1 | | Authors: | Ding, Z.Y, Huang, H.J. | | Deposit date: | 2022-10-09 | | Release date: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Cryo-EM structures reveal the activation and substrate recognition mechanism of human enteropeptidase.
Nat Commun, 13, 2022
|
|
7XAA
 
 | | Crystal structure of PDE4D catalytic domain complexed with compound 21d | | Descriptor: | 8-methoxy-2,2-dimethyl-7-(3-methylbut-2-enyl)-9-oxidanyl-5-(pyridin-4-ylmethoxy)pyrano[3,2-b]xanthen-6-one, Isoform 3 of cAMP-specific 3',5'-cyclic phosphodiesterase 4D, MAGNESIUM ION, ... | | Authors: | Huang, Y.-Y, He, X, Luo, H.-B. | | Deposit date: | 2022-03-17 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.100414 Å) | | Cite: | Discovery of novel PDE4 inhibitors targeting the M-pocket from natural mangostanin with improved safety for the treatment of Inflammatory Bowel Diseases.
Eur.J.Med.Chem., 242, 2022
|
|
6PFC
 
 | | Crystal structure of TS-DHFR from Cryptosporidium hominis in complex with NADPH, FdUMP and 2-(2-(4-((2-amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-5-yl)methyl)benzamido)-4-methoxyphenyl)acetic acid | | Descriptor: | 5-FLUORO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, Bifunctional dihydrofolate reductase-thymidylate synthase, METHOTREXATE, ... | | Authors: | Czyzyk, D.J, Valhondo, M, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2019-06-21 | | Release date: | 2019-10-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.689 Å) | | Cite: | Structure activity relationship towards design of cryptosporidium specific thymidylate synthase inhibitors.
Eur.J.Med.Chem., 183, 2019
|
|
3X2W
 
 | | Michaelis complex of cAMP-dependent Protein Kinase Catalytic Subunit | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CARBONATE ION, MAGNESIUM ION, ... | | Authors: | Das, A, Langan, P, Gerlits, O, Kovalevsky, A.Y, Heller, W.T. | | Deposit date: | 2015-01-02 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Protein Kinase A Catalytic Subunit Primed for Action: Time-Lapse Crystallography of Michaelis Complex Formation.
Structure, 23, 2015
|
|
6PFF
 
 | | Crystal structure of TS-DHFR from Cryptosporidium hominis in complex with NADPH, FdUMP and 2-(4-((2-amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-5-yl)methyl)benzamido)-4-cyanobenzoic acid. | | Descriptor: | 2-({4-[(2-amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-5-yl)methyl]benzene-1-carbonyl}amino)-4-cyanobenzoic acid, 5-FLUORO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | Authors: | Czyzyk, D.J, Valhondo, M, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2019-06-21 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.982 Å) | | Cite: | Structure activity relationship towards design of cryptosporidium specific thymidylate synthase inhibitors.
Eur.J.Med.Chem., 183, 2019
|
|
5MYV
 
 | | Crystal structure of SRPK2 in complex with compound 1 | | Descriptor: | 5-methyl-~{N}-[2-(4-methylpiperazin-1-yl)-5-(trifluoromethyl)phenyl]furan-2-carboxamide, DIMETHYL SULFOXIDE, SRSF protein kinase 2,SRSF protein kinase 2, ... | | Authors: | Chaikuad, A, Pike, A.C.W, Savitsky, P, von Delft, F, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-01-29 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Development of Potent, Selective SRPK1 Inhibitors as Potential Topical Therapeutics for Neovascular Eye Disease.
ACS Chem. Biol., 12, 2017
|
|
3X2V
 
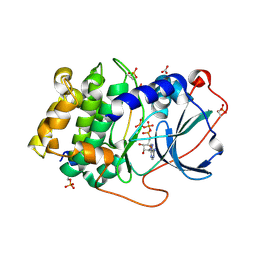 | | Michaelis-like complex of cAMP-dependent Protein Kinase Catalytic Subunit | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CARBONATE ION, MAGNESIUM ION, ... | | Authors: | Das, A, Langan, P, Gerlits, O, Kovalevsky, A.Y, Heller, W.T. | | Deposit date: | 2015-01-02 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Protein Kinase A Catalytic Subunit Primed for Action: Time-Lapse Crystallography of Michaelis Complex Formation.
Structure, 23, 2015
|
|
6W3T
 
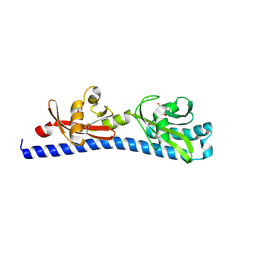 | | Crystal structure of ligand-binding domain of Campylobacter jejuni chemoreceptor Tlp3 in complex with L-norvaline | | Descriptor: | GLYCEROL, Methyl-accepting chemotaxis protein, NORVALINE, ... | | Authors: | Khan, M.F, Machuca, M.A, Rahman, M.M, Roujeinikova, A. | | Deposit date: | 2020-03-09 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Activity Relationship Study Reveals the Molecular Basis for Specific Sensing of Hydrophobic Amino Acids by theCampylobacter jejuniChemoreceptor Tlp3.
Biomolecules, 10, 2020
|
|
6WAT
 
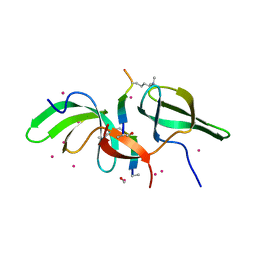 | | complex structure of PHF1 | | Descriptor: | Histone H3.1t peptide, PHD finger protein 1, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Min, J.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-03-26 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for histone variant H3tK27me3 recognition by PHF1 and PHF19.
Elife, 9, 2020
|
|
3WZJ
 
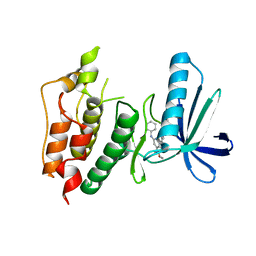 | | CRYSTAL STRUCTURE OF HUMAN MPS1 CATALYTIC DOMAIN IN COMPLEX WITH 4-(6-(cyclohexylamino)-8-(((tetrahydro-2H-pyran-4-yl)methyl)amino)imidazo[1,2-b]pyridazin-3-yl)-N-cyclopropylbenzamide | | Descriptor: | 4-{6-(cyclohexylamino)-8-[(tetrahydro-2H-pyran-4-ylmethyl)amino]imidazo[1,2-b]pyridazin-3-yl}-N-cyclopropylbenzamide, Dual specificity protein kinase TTK | | Authors: | Kusakabe, K, Ide, N, Daigo, Y, Itoh, T, Yamamoto, T, Kojima, E, Mitsuoka, Y, Tadano, G, Tagashira, S, Higashino, K, Okano, Y, Sato, Y, Inoue, M, Iguchi, M, Kanazawa, T, Ishioka, Y, Dohi, K, Kido, Y, Sakamoto, S, Ando, S, Maeda, M, Higaki, M, Yoshizawa, H, Mura, H, Nakamura, Y. | | Deposit date: | 2014-09-29 | | Release date: | 2015-02-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Discovery of imidazo[1,2-b]pyridazine derivatives: selective and orally available Mps1 (TTK) kinase inhibitors exhibiting remarkable antiproliferative activity.
J.Med.Chem., 58, 2015
|
|
8H3X
 
 | | Bacteroide Fragilis Toxin in complex with nanobody 282 | | Descriptor: | Fragilysin, ZINC ION, nanobody 282 | | Authors: | Wen, Y, Guo, Y. | | Deposit date: | 2022-10-09 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Screening and epitope characterization of diagnostic nanobody against total and activated Bacteroides fragilis toxin.
Front Immunol, 14, 2023
|
|
8H8J
 
 | | Lodoxamide-bound GPR35 in complex with G13 | | Descriptor: | 2,2'-[(2-chloro-5-cyano-1,3-phenylene)bis(azanediyl)]bis(oxoacetic acid), CALCIUM ION, CHOLESTEROL, ... | | Authors: | Yuan, Q, Duan, J, Liu, Q, Xu, H.E, Jiang, Y. | | Deposit date: | 2022-10-23 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Insights into divalent cation regulation and G 13 -coupling of orphan receptor GPR35.
Cell Discov, 8, 2022
|
|
8H3Y
 
 | | Bacteroide Fragilis Toxin in complex with nanobody 327 | | Descriptor: | Fragilysin, Nanobody 327, ZINC ION | | Authors: | Wen, Y, Guo, Y. | | Deposit date: | 2022-10-09 | | Release date: | 2023-02-08 | | Last modified: | 2023-03-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Screening and epitope characterization of diagnostic nanobody against total and activated Bacteroides fragilis toxin.
Front Immunol, 14, 2023
|
|
8HS3
 
 | | Gi bound orphan GPR20 in ligand-free state | | Descriptor: | Ggama, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Lin, X, Jiang, S, Xu, F. | | Deposit date: | 2022-12-16 | | Release date: | 2023-03-08 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | The activation mechanism and antibody binding mode for orphan GPR20.
Cell Discov, 9, 2023
|
|
8HSC
 
 | | Gi bound Orphan GPR20 complex with Fab046 in ligand-free state | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Lin, X, Jiang, S, Xu, F. | | Deposit date: | 2022-12-19 | | Release date: | 2023-03-08 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | The activation mechanism and antibody binding mode for orphan GPR20.
Cell Discov, 9, 2023
|
|
7XJW
 
 | | Crystal structure of canine coronavirus main protease in complex with GC376 | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, ORF1a polyprotein | | Authors: | Wang, Y.C, Yang, C.S, Hou, M.H, Tsai, C.L, Chiu, Y.F, Chen, Y. | | Deposit date: | 2022-04-18 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A Structural Comparison of SARS-CoV-2 Main Protease and Animal Coronaviral Main Protease Reveals Species-Specific Ligand Binding and Dimerization Mechanism.
Int J Mol Sci, 23, 2022
|
|
5N7F
 
 | | MAGI-1 complexed with a pRSK1 peptide | | Descriptor: | CALCIUM ION, GLYCEROL, Membrane-associated guanylate kinase, ... | | Authors: | Gogl, G, Nyitray, L. | | Deposit date: | 2017-02-20 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dynamic control of RSK complexes by phosphoswitch-based regulation.
FEBS J., 285, 2018
|
|
6Z2G
 
 | | Crystal structure of human NLRP9 PYD | | Descriptor: | NACHT, LRR and PYD domains-containing protein 9 | | Authors: | Marleaux, M, Anand, K, Geyer, M. | | Deposit date: | 2020-05-15 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the human NLRP9 pyrin domain suggests a distinct mode of inflammasome assembly.
Febs Lett., 594, 2020
|
|
6Z8L
 
 | | Alpha-Amylase in complex with probe fragments | | Descriptor: | CALCIUM ION, CHLORIDE ION, Pancreatic alpha-amylase, ... | | Authors: | Adam, S, Koehnke, J. | | Deposit date: | 2020-06-02 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.40000367 Å) | | Cite: | Enhancing glycan stability via site-selective fluorination: modulating substrate orientation by molecular design.
Chem Sci, 12, 2020
|
|
7BHD
 
 | | FimH in complex with alpha1,6 core-fucosylated oligomannose-3, crystallized in the trigonal space group | | Descriptor: | NICKEL (II) ION, SULFATE ION, Type 1 fimbrin D-mannose specific adhesin, ... | | Authors: | Bridot, C, Bouckaert, J, Krammer, E.-M. | | Deposit date: | 2021-01-11 | | Release date: | 2022-07-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural insights into a cooperative switch between one and two FimH bacterial adhesins binding pauci- and high-mannose type N-glycan receptors.
J.Biol.Chem., 299, 2023
|
|
3X2U
 
 | | Michaelis-like initial complex of cAMP-dependent Protein Kinase Catalytic Subunit. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Substrate Peptide, ... | | Authors: | Das, A, Langan, P, Gerlits, O, Kovalevsky, A.Y, Heller, W.T. | | Deposit date: | 2015-01-02 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Protein Kinase A Catalytic Subunit Primed for Action: Time-Lapse Crystallography of Michaelis Complex Formation.
Structure, 23, 2015
|
|
6ZJ9
 
 | | Crystal structure of Equus ferus caballus glutathione transferase A3-3 in complex with glutathione | | Descriptor: | 1,2-ETHANEDIOL, GLUTATHIONE, Glutathione S-transferase | | Authors: | Skerlova, J, Ismail, A, Lindstrom, H, Sjodin, B, Mannervik, B, Stenmark, P. | | Deposit date: | 2020-06-28 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional analysis of the inhibition of equine glutathione transferase A3-3 by organotin endocrine disrupting pollutants.
Environ Pollut, 268, 2021
|
|
