1HCA
 
 | |
1HCB
 
 | |
1HCC
 
 | |
1HCD
 
 | | STRUCTURE OF HISACTOPHILIN IS SIMILAR TO INTERLEUKIN-1 BETA AND FIBROBLAST GROWTH FACTOR | | Descriptor: | HISACTOPHILIN | | Authors: | Habazettl, J, Gondol, D, Wiltscheck, R, Otlewski, J, Schleicher, M, Holak, T.A. | | Deposit date: | 1994-05-03 | | Release date: | 1994-10-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of hisactophilin is similar to interleukin-1 beta and fibroblast growth factor.
Nature, 359, 1992
|
|
1HCE
 
 | | STRUCTURE OF HISACTOPHILIN IS SIMILAR TO INTERLEUKIN-1 BETA AND FIBROBLAST GROWTH FACTOR | | Descriptor: | HISACTOPHILIN | | Authors: | Habazettl, J, Gondol, D, Wiltscheck, R, Otlewski, J, Schleicher, M, Holak, T.A. | | Deposit date: | 1994-07-12 | | Release date: | 1994-09-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of hisactophilin is similar to interleukin-1 beta and fibroblast growth factor.
Nature, 359, 1992
|
|
1HCF
 
 | | Crystal structure of TrkB-d5 bound to neurotrophin-4/5 | | Descriptor: | BDNF/NT-3 GROWTH FACTORS RECEPTOR, NEUROTROPHIN-4, SULFATE ION | | Authors: | Banfield, M.J, Naylor, R.L, Robertson, A.G.S, Allen, S.J, Dawbarn, D, Brady, R.L. | | Deposit date: | 2001-05-03 | | Release date: | 2001-12-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Specificity in Trk-Receptor:Neurotrophin Interaction: The Crystal Structure of Trkb-D5 in Complex with Neurotrophin-4/5
Structure, 9, 2001
|
|
1HCG
 
 | |
1HCH
 
 | | Structure of horseradish peroxidase C1A compound I | | Descriptor: | ACETATE ION, CALCIUM ION, OXYGEN ATOM, ... | | Authors: | Berglund, G.I, Carlsson, G.H, Hajdu, J, Smith, A.T, Szoke, H, Henriksen, A. | | Deposit date: | 2001-05-04 | | Release date: | 2002-07-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | The Catalytic Pathway of Horseradish Peroxidase at High Resolution
Nature, 417, 2002
|
|
1HCI
 
 | |
1HCJ
 
 | |
1HCK
 
 | | HUMAN CYCLIN-DEPENDENT KINASE 2 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, HUMAN CYCLIN-DEPENDENT KINASE 2, MAGNESIUM ION | | Authors: | Schulze-Gahmen, U, De Bondt, H.L, Kim, S.-H. | | Deposit date: | 1996-06-03 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution crystal structures of human cyclin-dependent kinase 2 with and without ATP: bound waters and natural ligand as guides for inhibitor design.
J.Med.Chem., 39, 1996
|
|
1HCL
 
 | | HUMAN CYCLIN-DEPENDENT KINASE 2 | | Descriptor: | HUMAN CYCLIN-DEPENDENT KINASE 2 | | Authors: | Schulze-Gahmen, U, De Bondt, H.L, Kim, S.-H. | | Deposit date: | 1996-06-03 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution crystal structures of human cyclin-dependent kinase 2 with and without ATP: bound waters and natural ligand as guides for inhibitor design.
J.Med.Chem., 39, 1996
|
|
1HCM
 
 | |
1HCN
 
 | | STRUCTURE OF HUMAN CHORIONIC GONADOTROPIN AT 2.6 ANGSTROMS RESOLUTION FROM MAD ANALYSIS OF THE SELENOMETHIONYL PROTEIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HUMAN CHORIONIC GONADOTROPIN | | Authors: | Wu, H, Lustbader, J.W, Liu, Y, Canfield, R.E, Hendrickson, W.A. | | Deposit date: | 1994-07-01 | | Release date: | 1994-09-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of human chorionic gonadotropin at 2.6 A resolution from MAD analysis of the selenomethionyl protein.
Structure, 2, 1994
|
|
1HCO
 
 | |
1HCP
 
 | |
1HCQ
 
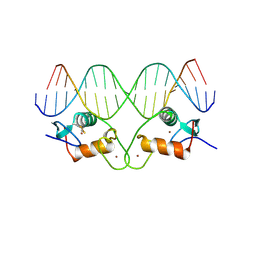 | | THE CRYSTAL STRUCTURE OF THE ESTROGEN RECEPTOR DNA-BINDING DOMAIN BOUND TO DNA: HOW RECEPTORS DISCRIMINATE BETWEEN THEIR RESPONSE ELEMENTS | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*GP*TP*CP*AP*CP*AP*GP*TP*GP*AP*CP*CP*T P*G)-3'), DNA (5'-D(*CP*CP*AP*GP*GP*TP*CP*AP*CP*TP*GP*TP*GP*AP*CP*CP*T P*G)-3'), PROTEIN (ESTROGEN RECEPTOR), ... | | Authors: | Schwabe, J.W.R, Chapman, L, Finch, J.T, Rhodes, D. | | Deposit date: | 1995-01-04 | | Release date: | 1995-11-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of the estrogen receptor DNA-binding domain bound to DNA: how receptors discriminate between their response elements.
Cell(Cambridge,Mass.), 75, 1993
|
|
1HCR
 
 | |
1HCS
 
 | |
1HCT
 
 | |
1HCU
 
 | | alpha-1,2-mannosidase from Trichoderma reesei | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-1,2-MANNOSIDASE, CALCIUM ION | | Authors: | Van Petegem, F, Contreras, H, Contreras, R, Van Beeumen, J. | | Deposit date: | 2001-05-09 | | Release date: | 2001-10-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Trichoderma Reesei Alpha-1,2-Mannosidase: Structural Basis for the Cleavage of Four Consecutive Mannose Residues
J.Mol.Biol., 312, 2001
|
|
1HCV
 
 | |
1HCW
 
 | |
1HCX
 
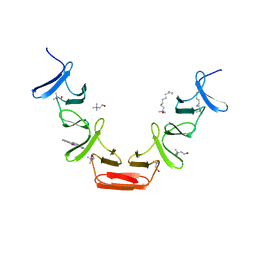 | | Choline binding domain of the major autolysin (C-LytA) from Streptococcus pneumoniae | | Descriptor: | 2,2':6',2''-TERPYRIDINE PLATINUM(II) Chloride, CHOLINE ION, DECYLAMINE-N,N-DIMETHYL-N-OXIDE, ... | | Authors: | Fernandez-Tornero, C, Lopez, R, Garcia, E, Gimenez-Gallego, G, Romero, A. | | Deposit date: | 2001-05-10 | | Release date: | 2001-11-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Novel Solenoid Fold in the Cell Wall Anchoring Domain of the Pneumococcal Virulence Factor Lyta
Nat.Struct.Biol., 8, 2001
|
|
1HCY
 
 | |
