4V42
 
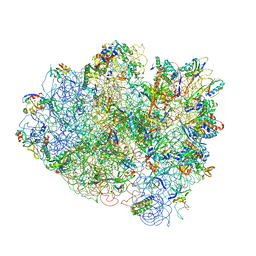 | | Crystal structure of the ribosome at 5.5 A resolution. | | Descriptor: | 30S 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Yusupov, M.M, Yusupova, G.Z, Baucom, A, Lieberman, K, Earnest, T.N, Cate, J.H.D, Noller, H.F. | | Deposit date: | 2001-03-30 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Crystal structure of the ribosome at 5.5 A resolution
Science, 292, 2001
|
|
4UPU
 
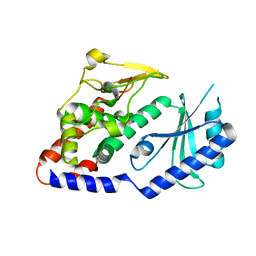
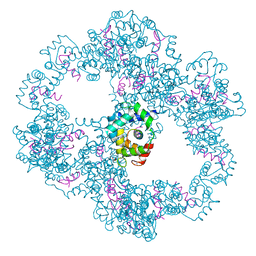 | | Crystal structure of IP3 3-K calmodulin binding region in complex with Calmodulin | | Descriptor: | CALCIUM ION, CALMODULIN, GLYCEROL, ... | | Authors: | Franco-Echevarria, E, Banos-Sanz, J.I, Monterroso, B, Round, A, Sanz-Aparicio, J, Gonzalez, B. | | Deposit date: | 2014-06-18 | | Release date: | 2014-08-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | A New Calmodulin Binding Motif for Inositol 1,4,5-Trisphosphate 3-Kinase Regulation.
Biochem.J., 463, 2014
|
|
4UD5
 
 | | Structural Plasticity of Cid1 Provides a Basis for its RNA Terminal Uridylyl Transferase Activity | | Descriptor: | POLY(A) RNA POLYMERASE PROTEIN CID1 | | Authors: | Yates, L.A, Durrant, B.P, Fleurdepine, S, Harlos, K, Norbury, C.J, Gilbert, R.J.C. | | Deposit date: | 2014-12-07 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structural Plasticity of Cid1 Provides a Basis for its Distributive RNA Terminal Uridylyl Transferase Activity.
Nucleic Acids Res., 43, 2015
|
|
9RAT
 
 | |
6EML
 
 | | Cryo-EM structure of a late pre-40S ribosomal subunit from Saccharomyces cerevisiae | | Descriptor: | 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, 40S ribosomal protein S11-A, ... | | Authors: | Heuer, A, Thomson, E, Schmidt, C, Berninghausen, O, Becker, T, Hurt, E, Beckmann, R. | | Deposit date: | 2017-10-02 | | Release date: | 2017-11-29 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of a late pre-40S ribosomal subunit fromSaccharomyces cerevisiae.
Elife, 6, 2017
|
|
4UP4
 
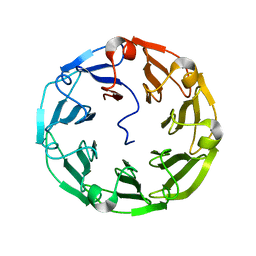 | | Structure of the recombinant lectin PVL from Psathyrella velutina in complex with GlcNAcb-D-1,3Galactoside | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose, ... | | Authors: | Audfray, A, Beljoudi, M, Hurbin, A, Varrot, A, Breiman, A, Busser, B, Lependu, J, Coll, J.L, Imberty, A. | | Deposit date: | 2014-06-12 | | Release date: | 2015-01-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Recombinant Fungal Lectin for Labeling Truncated Glycans on Human Cancer Cells.
Plos One, 10, 2015
|
|
6FAI
 
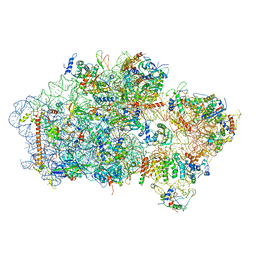
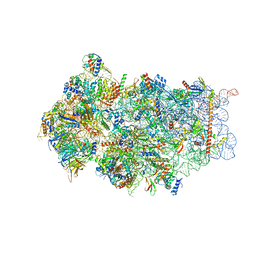 | | Structure of a eukaryotic cytoplasmic pre-40S ribosomal subunit | | Descriptor: | 20S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Scaiola, A, Pena, C, Weisser, M, Boehringer, D, Leibundgut, M, Klingauf-Nerurkar, P, Gerhardy, S, Panse, V.G, Ban, N. | | Deposit date: | 2017-12-15 | | Release date: | 2018-02-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of a eukaryotic cytoplasmic pre-40S ribosomal subunit.
EMBO J., 37, 2018
|
|
4UD4
 
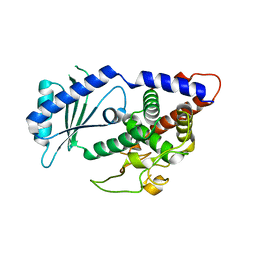 | | Structural Plasticity of Cid1 Provides a Basis for its RNA Terminal Uridylyl Transferase Activity | | Descriptor: | GLYCEROL, POLY(A) RNA POLYMERASE PROTEIN CID1 | | Authors: | Yates, L.A, Durrant, B.P, Fleurdepine, S, Harlos, K, Norbury, C.J, Gilbert, R.J.C. | | Deposit date: | 2014-12-07 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural plasticity of Cid1 provides a basis for its distributive RNA terminal uridylyl transferase activity.
Nucleic Acids Res., 43, 2015
|
|
9NTN
 
 | | Structure of Cap10-CdnD complex containing NDG modification | | Descriptor: | 2-amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidine-5-carboxylic acid, COENZYME A, Cap10, ... | | Authors: | Wassarman, D.R, Pfaff, P, Paulo, J.A, Gygi, S.P, Shokat, K.M, Kranzusch, P.J. | | Deposit date: | 2025-03-18 | | Release date: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Deazaguanylation is a nucleobase-protein conjugation required for type IV CBASS immunity.
Biorxiv, 2025
|
|
8BDZ
 
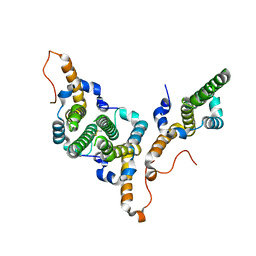 | | Hepatitis B virus core antigen (HBc) with the insertion of four external domains of the influenza A M2 protein (HBc/4M2e) with T=4 topology | | Descriptor: | Core protein,Matrix protein 2,External core antigen | | Authors: | Egorov, V.V, Shvetsov, A.V, Pichkur, E.B, Shaldzhyan, A.A, Zabrodskaya, Y.A, Vinogradova, D.S, Nekrasov, P.A, Gorshkov, A.N, Garmay, Y.P, Kovaleva, A.A, Stepanova, L.A, Tsybalova, L.M, Shtam, T.A, Myasnikov, A.G, Konevega, A.L. | | Deposit date: | 2022-10-20 | | Release date: | 2022-12-28 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Inside and outside of virus-like particles HBc and HBc/4M2e: A comprehensive study of the structure.
Biophys.Chem., 293, 2022
|
|
8BER
 
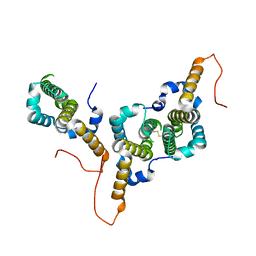 | | Hepatitis B virus core antigen (HBc) with the insertion of four external domains of the influenza A M2 protein (HBc/4M2e) with T=3 topology | | Descriptor: | Core protein,Matrix protein 2,External core antigen | | Authors: | Egorov, V.V, Shvetsov, A.V, Pichkur, E.B, Shaldzhyan, A.A, Zabrodskaya, Y.A, Vinogradova, D.S, Nekrasov, P.A, Gorshkov, A.N, Garmay, Y.P, Kovaleva, A.A, Stepanova, L.A, Tsybalova, L.M, Shtam, T.A, Myasnikov, A.G, Konevega, A.L. | | Deposit date: | 2022-10-21 | | Release date: | 2022-12-28 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Inside and outside of virus-like particles HBc and HBc/4M2e: A comprehensive study of the structure.
Biophys.Chem., 293, 2022
|
|
6M8Z
 
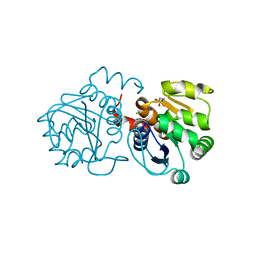 | | Crystal structure of human DJ-1 without a modification on Cys-106 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, Protein/nucleic acid deglycase DJ-1 | | Authors: | Shumilin, I.A, Shabalin, I.G, Shumilina, S.V, Werenskjold, C, Utepbergenov, D, Minor, W. | | Deposit date: | 2018-08-22 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | A transient post-translational modification of active site cysteine alters binding properties of the parkinsonism protein DJ-1.
Biochem. Biophys. Res. Commun., 504, 2018
|
|
9LK5
 
 | | The structure of Lhcb8-C2S2 PSII-LHCII supercomplex from Arabidopsis thaliana | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Zhou, Q, Caferri, R, Shan, J.Y, Amelii, A, Bassi, R, Liu, Z.F. | | Deposit date: | 2025-01-16 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A stress-induced paralog of Lhcb4 controls the photosystem II functional architecture in Arabidopsis thaliana
To Be Published
|
|
9LK4
 
 | | Cryo-EM structure of Lhcb4.1-C2S2 PSII-LHCII supercomplex from Arabidopsis thaliana | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Zhou, Q, Caferri, R, Shan, J.Y, Amelii, A, Bassi, R, Liu, Z.F. | | Deposit date: | 2025-01-16 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A stress-induced paralog of Lhcb4 controls the photosystem II functional architecture in Arabidopsis thaliana
To Be Published
|
|
9NR5
 
 | | Crystal structure of H5 hemagglutinin Q226L mutant from the influenza virus A/black swan/Akita/1/2016 with LSTc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, ... | | Authors: | Lin, T.H, Zhu, Y, Wilson, I.A. | | Deposit date: | 2025-03-14 | | Release date: | 2025-04-23 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | The Q226L mutation can convert a highly pathogenic H5 2.3.4.4e virus to bind human-type receptors.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9NRB
 
 | | Crystal structure of H5 hemagglutinin Q226L mutant from the influenza virus A/duck/France/1611008h/16 with LSTc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, ... | | Authors: | Lin, T.H, Zhu, Y, Wilson, I.A. | | Deposit date: | 2025-03-14 | | Release date: | 2025-04-23 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The Q226L mutation can convert a highly pathogenic H5 2.3.4.4e virus to bind human-type receptors.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9NR2
 
 | | Crystal structure of H5 hemagglutinin from the influenza virus A/black swan/Akita/1/2016 with LSTa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, Hemagglutinin HA2, ... | | Authors: | Lin, T.H, Zhu, Y, Wilson, I.A. | | Deposit date: | 2025-03-13 | | Release date: | 2025-04-23 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | The Q226L mutation can convert a highly pathogenic H5 2.3.4.4e virus to bind human-type receptors.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9RSA
 
 | |
7NSN
 
 | | Multi-domain GH92 alpha-1,2-mannosidase from Neobacillus novalis: mannoimidazole complex | | Descriptor: | (5R,6R,7S,8R)-5-(HYDROXYMETHYL)-5,6,7,8-TETRAHYDROIMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Kolaczkowski, B.M, Moroz, O.V, Blagova, E, Davies, G.J, Wilson, K.S, Moeler, M.S, Meyer, A.S, Westh, P, Jensen, K, Krogh, K.B.R.M. | | Deposit date: | 2021-03-08 | | Release date: | 2022-09-21 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural and functional characterization of a multi-domain GH92 alpha-1,2-mannosidase from Neobacillus novalis.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7OCV
 
 | | Human TNKS1 in complex with 3-[4-(1-Hydroxy-1-methyl-ethyl)-phenyl]-6-methyl-2H-pyrrolo[1,2-a]pyrazin-1-one | | Descriptor: | 6-methyl-3-[4-(2-oxidanylpropan-2-yl)phenyl]-4~{H}-pyrrolo[1,2-a]pyrazin-1-one, ACETATE ION, Poly [ADP-ribose] polymerase, ... | | Authors: | Musil, D, Lehmann, M, Buchstaller, H.-P. | | Deposit date: | 2021-04-28 | | Release date: | 2021-07-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.432 Å) | | Cite: | Optimization of a Screening Hit toward M2912, an Oral Tankyrase Inhibitor with Antitumor Activity in Colorectal Cancer Models.
J.Med.Chem., 64, 2021
|
|
7NJC
 
 | |
6ZUZ
 
 | |
7POG
 
 | | High-resolution structure of native toxin A from Clostridioides difficile | | Descriptor: | Toxin A, ZINC ION | | Authors: | Boesen, T, Joergensen, R, Aminzadeh, A, Engelbrecht Larsen, C. | | Deposit date: | 2021-09-08 | | Release date: | 2021-12-08 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | High-resolution structure of native toxin A from Clostridioides difficile.
Embo Rep., 23, 2022
|
|
7OBN
 
 | | Structural investigations of a new L3 DNA ligase: structure-function analysis | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA (5'-D(*TP*TP*CP*CP*GP*AP*TP*AP*GP*TP*GP*GP*GP*GP*TP*CP*GP*CP*AP*AP*T)-3'), DNA ligase, ... | | Authors: | Leiros, H.-K.S, Williamson, A. | | Deposit date: | 2021-04-23 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Bacteriophage origin of some minimal ATP-dependent DNA ligases: a new structure from Burkholderia pseudomallei with striking similarity to Chlorella virus ligase.
Sci Rep, 11, 2021
|
|
6G8J
 
 | | 14-3-3sigma in complex with a A130beta3A mutated YAP pS127 phosphopeptide | | Descriptor: | 14-3-3 protein sigma, ACE-ARG-ALA-HIS-SEP-SER-PRO-BAL-SER-LEU-GLN, CHLORIDE ION, ... | | Authors: | Andrei, S.A, Thijssen, V, Brunsveld, L, Ottmann, C, Milroy, L.G. | | Deposit date: | 2018-04-09 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | A study on the effect of synthetic alpha-to-beta3-amino acid mutations on the binding of phosphopeptides to 14-3-3 proteins.
Chem.Commun.(Camb.), 55, 2019
|
|
