1CY6
 
 | |
4AN8
 
 | |
1CY1
 
 | | COMPLEX OF E.COLI DNA TOPOISOMERASE I WITH 5'PTPTPT | | Descriptor: | DNA TOPOISOMERASE I, PHOSPHATE ION, THYMIDINE-3',5'-DIPHOSPHATE, ... | | Authors: | Feinberg, H, Changela, A, Mondragon, A. | | Deposit date: | 1999-08-31 | | Release date: | 2000-03-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Protein-nucleotide interactions in E. coli DNA topoisomerase I.
Nat.Struct.Biol., 6, 1999
|
|
1CY9
 
 | |
229D
 
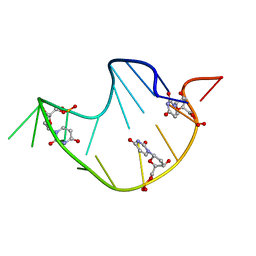 | | DNA ANALOG OF YEAST TRANSFER RNA PHE ANTICODON DOMAIN WITH MODIFIED BASES 5-METHYL CYTOSINE AND 1-METHYL GUANINE | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*AP*CP*(UMP)P*GP*AP*AP*(MG1)P*AP*(UMP)P*(5CM)P*(UMP)P*GP*G)-3') | | Authors: | Basti, M.M, Stuart, J.W, Lam, A.T, Guenther, R, Agris, P.F. | | Deposit date: | 1995-08-16 | | Release date: | 1995-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Design, biological activity and NMR-solution structure of a DNA analogue of yeast tRNA(Phe) anticodon domain.
Nat.Struct.Biol., 3, 1996
|
|
1CY2
 
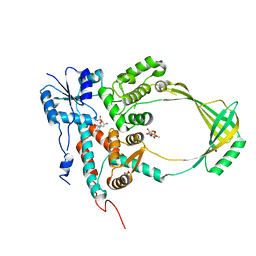 | |
1CY8
 
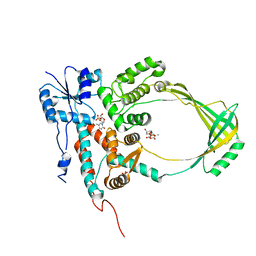 | | COMPLEX OF E.COLI DNA TOPOISOMERASE I WITH 5'-THYMIDINE MONOPHOSPHATE AND 3'-THYMIDINE MONOPHOSPHATE | | Descriptor: | DNA TOPOISOMERASE I, PHOSPHATE ION, THYMIDINE-3'-PHOSPHATE, ... | | Authors: | Feinberg, H, Changela, A, Mondragon, A. | | Deposit date: | 1999-08-31 | | Release date: | 2000-03-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Protein-nucleotide interactions in E. coli DNA topoisomerase I.
Nat.Struct.Biol., 6, 1999
|
|
1CYY
 
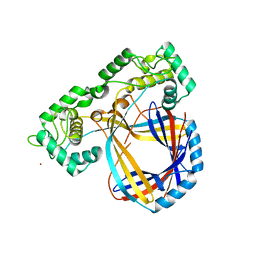 | |
3GOD
 
 | |
5TRN
 
 | | Solution Structure of a DNA Dodecamer with 8-oxoguanine at the 4th position and 5-methylcytosine at the 9th position | | Descriptor: | DNA (5'-D(*CP*GP*CP*(8OG)P*AP*AP*TP*TP*(DMC)P*GP*CP*G)-3') | | Authors: | Hoppins, J.J, Gruber, D.R, Miears, H.L, Endutkin, A.V, Zharkov, D.O, Smirnov, S.L. | | Deposit date: | 2016-10-26 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Oxidative damage to epigenetically methylated sites affects DNA stability, dynamics and enzymatic demethylation.
Nucleic Acids Res., 46, 2018
|
|
5ZB2
 
 | | Crystal structure of Rad7 and Elc1 complex in yeast | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 3,6,9,12,15,18,21,24,27,30,33,36,39,42,45,48,51,54,57-nonadecaoxanonapentacontane-1,59-diol, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ... | | Authors: | Jiang, T, Liu, L, Huo, Y. | | Deposit date: | 2018-02-09 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.30000544 Å) | | Cite: | Crystal structure of the yeast Rad7-Elc1 complex and assembly of the Rad7-Rad16-Elc1-Cul3 complex.
DNA Repair (Amst.), 77, 2019
|
|
7D9K
 
 | |
7D9Y
 
 | |
5VI0
 
 | |
5Y7Q
 
 | | Crystal structure of paFAN1 bound to 2nt 5'flap DNA with gap | | Descriptor: | DNA (5'-D(*TP*TP*CP*AP*CP*AP*CP*AP*TP*TP*CP*AP*A)-3'), DNA (5'-D(P*GP*AP*AP*TP*GP*TP*GP*TP*GP*TP*CP*TP*CP*AP*AP*TP*CP*CP*CP*AP*AP*CP*TP*T)-3'), DNA (5'-D(P*GP*TP*TP*GP*GP*GP*AP*TP*TP*G)-3'), ... | | Authors: | Jin, H, Cho, Y. | | Deposit date: | 2017-08-17 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural mechanism of DNA interstrand cross-link unhooking by the bacterial FAN1 nuclease.
J. Biol. Chem., 293, 2018
|
|
7E4E
 
 | |
5Z6W
 
 | | Crystal structure of paFAN1 bound to 2nt 5'flap DNA with gap with Manganese | | Descriptor: | DNA (5'-D(P*AP*TP*TP*CP*AP*A)-3'), DNA (5'-D(P*GP*AP*AP*TP*GP*TP*GP*TP*CP*TP*CP*AP*AP*TP*CP*CP*CP*AP*AP*CP*TP*T)-3'), DNA (5'-D(P*GP*TP*TP*GP*GP*GP*AP*TP*TP*G)-3'), ... | | Authors: | Jin, H, Cho, Y. | | Deposit date: | 2018-01-25 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural mechanism of DNA interstrand cross-link unhooking by the bacterial FAN1 nuclease.
J. Biol. Chem., 293, 2018
|
|
1Z8S
 
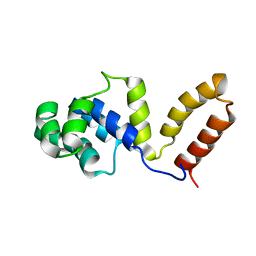 | | DnaB binding domain of DnaG (P16) from Bacillus stearothermophilus (residues 452-597) | | Descriptor: | DNA primase | | Authors: | Syson, K, Thirlway, J, Hounslow, A.M, Soultanas, P, Waltho, J.P. | | Deposit date: | 2005-03-31 | | Release date: | 2005-10-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the helicase-interaction domain of the primase DnaG: a model for helicase activation
Structure, 13, 2005
|
|
6A5N
 
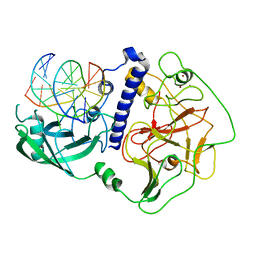 | | Crystal structure of Arabidopsis thaliana SUVH6 in complex with methylated DNA | | Descriptor: | DNA (5'-D(*CP*AP*CP*TP*GP*CP*TP*GP*AP*GP*TP*AP*CP*T)-3'), DNA (5'-D(*GP*AP*GP*TP*AP*CP*TP*(5CM)P*AP*GP*CP*AP*GP*T)-3'), Histone-lysine N-methyltransferase, ... | | Authors: | Li, X, Du, J. | | Deposit date: | 2018-06-24 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanistic insights into plant SUVH family H3K9 methyltransferases and their binding to context-biased non-CG DNA methylation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6KDB
 
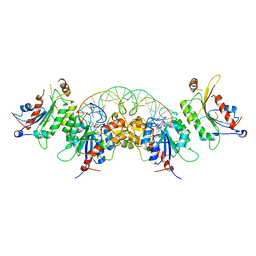 | | Crystal structure of human DNMT3B-DNMT3L in complex with DNA containing CpGpT site | | Descriptor: | DNA (25-MER), DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3B, ... | | Authors: | Lin, C.-C, Chen, Y.-P, Yang, W.-Z, Shen, C.-K, Yuan, H.S. | | Deposit date: | 2019-07-01 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.862 Å) | | Cite: | Structural insights into CpG-specific DNA methylation by human DNA methyltransferase 3B.
Nucleic Acids Res., 48, 2020
|
|
6KDA
 
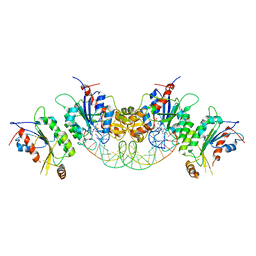 | | Crystal structure of human DNMT3B-DNMT3L in complex with DNA containing CpGpG site | | Descriptor: | DNA (25-MER), DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3B, ... | | Authors: | Lin, C.-C, Chen, Y.-P, Yang, W.-Z, Shen, C.-K, Yuan, H.S. | | Deposit date: | 2019-07-01 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.909 Å) | | Cite: | Structural insights into CpG-specific DNA methylation by human DNA methyltransferase 3B.
Nucleic Acids Res., 48, 2020
|
|
286D
 
 | |
285D
 
 | |
8POP
 
 | | HK97 small terminase in complex with DNA | | Descriptor: | DNA (31-MER), Terminase small subunit | | Authors: | Chechik, M, Greive, S.J, Antson, A.A, Jenkins, H.T. | | Deposit date: | 2023-07-05 | | Release date: | 2023-08-09 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for DNA recognition by a viral genome-packaging machine.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6WJK
 
 | | Crystal Structure of a Self-Assembling DNA Crystal Scaffold with Rhombohedral Symmetry | | Descriptor: | DNA (32-MER), DNA (5'-D(*TP*GP*GP*AP*AP*AP*CP*AP*GP*AP*CP*TP*GP*TP*CP*AP*GP*AP*TP*G)-3'), DNA (5'-D(P*AP*GP*CP*AP*TP*GP*A)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-04-13 | | Release date: | 2021-02-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (4.514 Å) | | Cite: | A Self-Assembled Rhombohedral DNA Crystal Scaffold with Tunable Cavity Sizes and High-Resolution Structural Detail.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
