5I6X
 
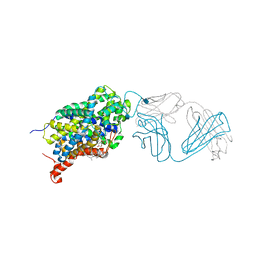 | | X-ray structure of the ts3 human serotonin transporter complexed with paroxetine at the central site | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 8B6 antibody, heavy chain, ... | | Authors: | Coleman, J.A, Green, E.M, Gouaux, E. | | Deposit date: | 2016-02-16 | | Release date: | 2016-04-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | X-ray structures and mechanism of the human serotonin transporter.
Nature, 532, 2016
|
|
1KUY
 
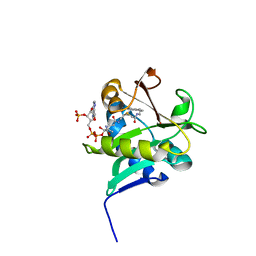 | | X-ray Crystallographic Studies of Serotonin N-acetyltransferase Catalysis and Inhibition | | Descriptor: | COA-S-ACETYL TRYPTAMINE, Serotonin N-acetyltransferase | | Authors: | Wolf, E, De Angelis, J, Khalil, E.M, Cole, P.A, Burley, S.K. | | Deposit date: | 2002-01-22 | | Release date: | 2002-03-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray crystallographic studies of serotonin N-acetyltransferase catalysis and inhibition.
J.Mol.Biol., 317, 2002
|
|
1KIK
 
 | | SH3 Domain of Lymphocyte Specific Kinase (LCK) | | Descriptor: | PROTO-ONCOGENE TYROSINE-PROTEIN KINASE LCK | | Authors: | Briese, L, Willbold, D. | | Deposit date: | 2001-12-03 | | Release date: | 2001-12-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure determination of human Lck unique and SH3 domains by nuclear magnetic resonance spectroscopy.
Bmc Struct.Biol., 3, 2003
|
|
1KMS
 
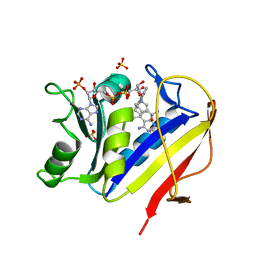 | | HUMAN DIHYDROFOLATE REDUCTASE COMPLEXED WITH NADPH AND 6-([5-QUINOLYLAMINO]METHYL)-2,4-DIAMINO-5-METHYLPYRIDO[2,3-D]PYRIMIDINE (SRI-9439), A LIPOPHILIC ANTIFOLATE | | Descriptor: | 6-([5-QUINOLYLAMINO]METHYL)-2,4-DIAMINO-5-METHYLPYRIDO[2,3-D]PYRIMIDINE, DIHYDROFOLATE REDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Klon, A.E, Heroux, A, Ross, L.J, Pathak, V, Johnson, C.A, Piper, J.R, Borhani, D.W. | | Deposit date: | 2001-12-17 | | Release date: | 2002-07-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Atomic structures of human dihydrofolate reductase complexed with NADPH and two lipophilic antifolates at 1.09 a and 1.05 a resolution.
J.Mol.Biol., 320, 2002
|
|
1KLD
 
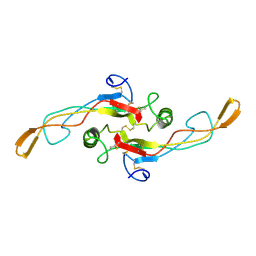 | | SOLUTION STRUCTURE OF TGF-B1, NMR, MODELS 18-33 OF 33 STRUCTURES | | Descriptor: | TRANSFORMING GROWTH FACTOR-BETA 1 | | Authors: | Hinck, A.P, Archer, S.J, Qian, S.W, Roberts, A.B, Sporn, M.B, Weatherbee, J.A, Tsang, M.L.-S, Lucas, R, Zhang, B.-L, Wenker, J, Torchia, D.A. | | Deposit date: | 1996-01-16 | | Release date: | 1996-08-17 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Transforming growth factor beta 1: three-dimensional structure in solution and comparison with the X-ray structure of transforming growth factor beta 2.
Biochemistry, 35, 1996
|
|
1KV4
 
 | | Solution structure of antibacterial peptide (Moricin) | | Descriptor: | moricin | | Authors: | Hemmi, H, Ishibashi, J, Hara, S, Yamakawa, M. | | Deposit date: | 2002-01-25 | | Release date: | 2002-05-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of moricin, an antibacterial peptide, isolated from the silkworm Bombyx mori.
FEBS Lett., 518, 2002
|
|
8F6F
 
 | | Cryo-EM structure of a Zinc-loaded D51A mutant of the YiiP-Fab complex | | Descriptor: | Cadmium and zinc efflux pump FieF, Fab2r heavy chain, Fab2r light chain, ... | | Authors: | Lopez-Redondo, M.L, Hussein, A.K, Stokes, D.L. | | Deposit date: | 2022-11-16 | | Release date: | 2023-02-08 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Energy coupling and stoichiometry of Zn 2+ /H + antiport by the prokaryotic cation diffusion facilitator YiiP.
Elife, 12, 2023
|
|
9FB2
 
 | | Gcase in complex with small molecule inhibitor 1 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Tisi, D, Cleasby, A. | | Deposit date: | 2024-05-11 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Fragment-Based Discovery of a Series of Allosteric-Binding Site Modulators of beta-Glucocerebrosidase.
J.Med.Chem., 67, 2024
|
|
4X5W
 
 | | HLA-DR1 with CLIP102-120(M107W) | | Descriptor: | HLA class II histocompatibility antigen gamma chain, HLA class II histocompatibility antigen, DR alpha chain, ... | | Authors: | Guenther, S, Freund, C. | | Deposit date: | 2014-12-06 | | Release date: | 2016-03-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | MHC class II complexes sample intermediate states along the peptide exchange pathway.
Nat Commun, 7, 2016
|
|
1KVV
 
 | | Solution Structure Of Protein SRP19 Of The Archaeoglobus fulgidus Signal Recognition Particle, Minimized Average Structure | | Descriptor: | SRP19 | | Authors: | Pakhomova, O.N, Deep, S, Huang, Q, Zwieb, C, Hinck, A.P. | | Deposit date: | 2002-01-27 | | Release date: | 2002-03-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of protein SRP19 of Archaeoglobus fulgidus signal recognition particle.
J.Mol.Biol., 317, 2002
|
|
1KW0
 
 | | Catalytic Domain of Human Phenylalanine Hydroxylase (Fe(II)) in Complex with Tetrahydrobiopterin and Thienylalanine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, BETA(2-THIENYL)ALANINE, FE (II) ION, ... | | Authors: | Andersen, O.A, Flatmark, T, Hough, E. | | Deposit date: | 2002-01-28 | | Release date: | 2003-01-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Ternary Complex of the Catalytic
Domain of Human Phenylalanine Hydroxylase with Tetrahydrobiopterin
and 3-(2-thienyl)-L-alanine, and its Implications for the Mechanism
of Catalysis and Substrate Activation
J.Mol.Biol., 320, 2002
|
|
1KWP
 
 | | Crystal Structure of MAPKAP2 | | Descriptor: | MAP Kinase Activated Protein Kinase 2, MERCURY (II) ION | | Authors: | Meng, W, Swenson, L.L, Fitzgibbon, M.J, Hayakawa, K, ter Haar, E, Behrens, A.E, Fulghum, J.R, Lippke, J.A. | | Deposit date: | 2002-01-30 | | Release date: | 2002-09-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Mitogen-activated Protein Kinase-activated Protein (MAPKAP) Kinase 2 Suggests a Bifunctional Switch That
Couples Kinase Activation with Nuclear Export
J.Biol.Chem., 277, 2002
|
|
5I1V
 
 | |
1KN1
 
 | | Crystal structure of allophycocyanin | | Descriptor: | Allophycocyanin, PHYCOCYANOBILIN | | Authors: | Liang, D.C, Liu, J.Y, Jiang, T, Zhang, J.P, Chang, W.R. | | Deposit date: | 2001-12-18 | | Release date: | 2002-12-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Allophycocyanin from red algae Porphyra yezoensis at 2.2 A resolution
J.BIOL.CHEM., 274, 1999
|
|
1KNA
 
 | |
1KNE
 
 | |
6UY5
 
 | |
1KXG
 
 | | The 2.0 Ang Resolution Structure of BLyS, B Lymphocyte Stimulator. | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, B lymphocyte stimulator, CITRIC ACID, ... | | Authors: | Oren, D.A, Li, Y, Volovik, Y, Morris, T.S, Dharia, C, Das, K, Galperina, O, Gentz, R, Arnold, E. | | Deposit date: | 2002-01-31 | | Release date: | 2002-03-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of BLyS receptor recognition.
Nat.Struct.Biol., 9, 2002
|
|
9FDI
 
 | | Gcase in complex with small molecule inhibitor 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(phenylmethyl)piperidine, ... | | Authors: | Tisi, D, Cleasby, A. | | Deposit date: | 2024-05-17 | | Release date: | 2024-07-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Fragment-Based Discovery of a Series of Allosteric-Binding Site Modulators of beta-Glucocerebrosidase.
J.Med.Chem., 67, 2024
|
|
1KXW
 
 | |
1KF5
 
 | | Atomic Resolution Structure of RNase A at pH 7.1 | | Descriptor: | pancreatic ribonuclease | | Authors: | Berisio, R, Sica, F, Lamzin, V.S, Wilson, K.S, Zagari, A, Mazzarella, L. | | Deposit date: | 2001-11-19 | | Release date: | 2001-12-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Atomic resolution structures of ribonuclease A at six pH values.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1KNG
 
 | | Crystal structure of CcmG reducing oxidoreductase at 1.14 A | | Descriptor: | THIOL:DISULFIDE INTERCHANGE PROTEIN CYCY | | Authors: | Edeling, M.A, Guddat, L.W, Fabianek, R.A, Thony-Meyer, L, Martin, J.L. | | Deposit date: | 2001-12-18 | | Release date: | 2002-07-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Structure of CcmG/DsbE at 1.14 A resolution: high-fidelity reducing activity in an indiscriminately oxidizing environment
Structure, 10, 2002
|
|
9F9Z
 
 | | Gcase in complex with small molecule inhibitor 1 | | Descriptor: | (2~{S})-1-(2,6-dimethylphenoxy)propan-2-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lysosomal acid glucosylceramidase, ... | | Authors: | Tisi, D, Cleasby, A. | | Deposit date: | 2024-05-09 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.279 Å) | | Cite: | Fragment-Based Discovery of a Series of Allosteric-Binding Site Modulators of beta-Glucocerebrosidase.
J.Med.Chem., 67, 2024
|
|
8EIU
 
 | | E. coli 70S ribosome with A-loop mutations U2554C and U2555C | | Descriptor: | 16S rRNA, 23S rRNA, 3'-amino-3'-deoxyadenosine 5'-(dihydrogen phosphate), ... | | Authors: | Nissley, A.J, Penev, P.I, Watson, Z.L, Banfield, J.F, Cate, J.H.D. | | Deposit date: | 2022-09-15 | | Release date: | 2023-02-01 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | Rare ribosomal RNA sequences from archaea stabilize the bacterial ribosome.
Nucleic Acids Res., 51, 2023
|
|
4X0T
 
 | |
