3FTZ
 
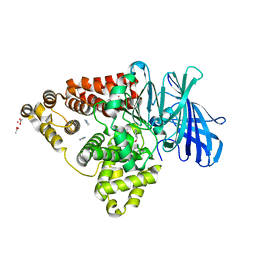 | | Leukotriene A4 hydrolase in complex with fragment 2-(pyridin-3-ylmethoxy)aniline | | Descriptor: | 2-(pyridin-3-ylmethoxy)aniline, ACETATE ION, IMIDAZOLE, ... | | Authors: | Davies, D.R. | | Deposit date: | 2009-01-13 | | Release date: | 2010-01-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of 4-[(2S)-2-{[4-(4-chlorophenoxy)phenoxy]methyl}-1-pyrrolidinyl]butanoic acid (DG-051) as a novel leukotriene A4 hydrolase inhibitor of leukotriene B4 biosynthesis.
J.Med.Chem., 53, 2010
|
|
3FTO
 
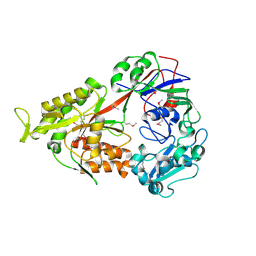 | | Crystal structure of OppA in a open conformation | | Descriptor: | Oligopeptide-binding protein oppA | | Authors: | Berntsson, R.P.-A, Oktaviani, N.A, Fusetti, F, Thunnissen, A.-M.W.H, Poolman, B, Slotboom, D.-J. | | Deposit date: | 2009-01-13 | | Release date: | 2009-03-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Selenomethionine incorporation in proteins expressed in Lactococcus lactis.
Protein Sci., 18, 2009
|
|
3FU5
 
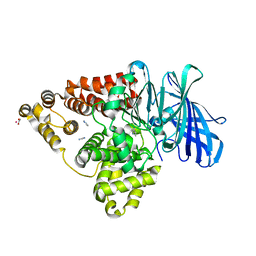 | |
3FUJ
 
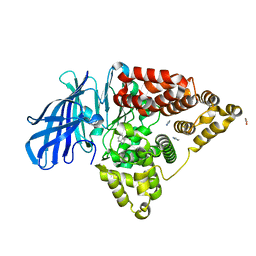 | |
1Z95
 
 | | Crystal Structure of the Androgen Receptor Ligand-binding Domain W741L Mutant Complex with R-bicalutamide | | Descriptor: | Androgen Receptor, R-BICALUTAMIDE, SULFATE ION | | Authors: | Bohl, C.E, Gao, W, Miller, D.D, Bell, C.E, Dalton, J.T. | | Deposit date: | 2005-03-31 | | Release date: | 2005-04-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for antagonism and resistance of bicalutamide in prostate cancer.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
3FTV
 
 | |
2BMK
 
 | |
2BSA
 
 | | Ferredoxin-Nadp Reductase (Mutation: Y 303 S) complexed with NADP | | Descriptor: | FERREDOXIN-NADP REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Maya, C.M, Hermoso, J.A, Perez-Dorado, I, Tejero, J, Julvez, M.M, Gomez-Moreno, C, Medina, M. | | Deposit date: | 2005-05-20 | | Release date: | 2005-10-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | C-Terminal Tyrosine of Ferredoxin-Nadp(+) Reductase in Hydride Transfer Processes with Nad(P)(+)/H.
Biochemistry, 44, 2005
|
|
3ZWH
 
 | | Ca2+-bound S100A4 C3S, C81S, C86S and F45W mutant complexed with myosin IIA | | Descriptor: | ACETATE ION, AZIDE ION, CALCIUM ION, ... | | Authors: | Kiss, B, Duelli, A, Radnai, L, Kekesi, A.K, Katona, G, Nyitray, L. | | Deposit date: | 2011-07-31 | | Release date: | 2012-04-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal Structure of the S100A4-Nonmuscle Myosin Iia Tail Fragment Complex Reveals an Asymmetric Target Binding Mechanism.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4C7D
 
 | | Structure and activity of the GH20 beta-N-acetylhexosaminidase from Streptomyces coelicolor A3(2) | | Descriptor: | 1,2-ETHANEDIOL, BETA-N-ACETYLHEXOSAMINIDASE | | Authors: | Nguyenthi, N, Offen, W.A, Davies, G.J, Doucet, N. | | Deposit date: | 2013-09-20 | | Release date: | 2014-03-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and Activity of the Streptomyces Coelicolor A3(2) Beta-N-Acetylhexosaminidase Provides Further Insight Into Gh20 Family Catalysis and Inhibition.
Biochemistry, 53, 2014
|
|
2AX9
 
 | | Crystal Structure Of The Androgen Receptor Ligand Binding Domain In Complex With R-3 | | Descriptor: | (R)-3-BROMO-2-HYDROXY-2-METHYL-N-[4-NITRO-3-(TRIFLUOROMETHYL)PHENYL]PROPANAMIDE, Androgen receptor | | Authors: | Bohl, C.E, Miller, D.D, Chen, J, Bell, C.E, Dalton, J.T. | | Deposit date: | 2005-09-03 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Basis for Accommodation of Nonsteroidal Ligands in the Androgen Receptor
J.Biol.Chem., 280, 2005
|
|
2C9T
 
 | | Crystal Structure Of Acetylcholine Binding Protein (AChBP) From Aplysia Californica In Complex With alpha-Conotoxin ImI | | Descriptor: | ALPHA-CONOTOXIN IMI, SOLUBLE ACETYLCHOLINE RECEPTOR | | Authors: | Ulens, C, Hogg, R.C, Celie, P.H, Bertrand, D, Tsetlin, V, Smit, A.B, Sixma, T.K. | | Deposit date: | 2005-12-14 | | Release date: | 2006-02-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Determinants of Selective {Alpha}-Conotoxin Binding to a Nicotinic Acetylcholine Receptor Homolog Achbp.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2CE9
 
 | |
2AFM
 
 | | Crystal structure of human glutaminyl cyclase at pH 6.5 | | Descriptor: | Glutaminyl-peptide cyclotransferase, SULFATE ION, ZINC ION | | Authors: | Huang, K.F, Liu, Y.L, Cheng, W.J, Ko, T.P, Wang, A.H.J. | | Deposit date: | 2005-07-26 | | Release date: | 2005-08-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structures of human glutaminyl cyclase, an enzyme responsible for protein N-terminal pyroglutamate formation
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1ZWD
 
 | |
2A8B
 
 | | Crystal Structure of the Catalytic Domain of Human Tyrosine Phosphatase Receptor, Type R | | Descriptor: | CHLORIDE ION, Receptor-type tyrosine-protein phosphatase R | | Authors: | Ugochukwu, E, Eswaran, J, Barr, A, Longman, E, Arrowsmith, C, Edwards, A, Sundstrom, M, von Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-07-07 | | Release date: | 2005-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures and inhibitor identification for PTPN5, PTPRR and PTPN7: a family of human MAPK-specific protein tyrosine phosphatases.
Biochem.J., 395, 2006
|
|
1Y58
 
 | |
2ADF
 
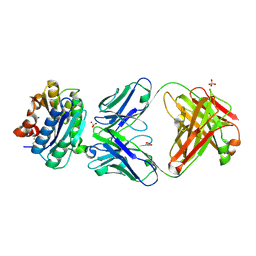 | | Crystal Structure and Paratope Determination of 82D6A3, an Antithrombotic Antibody Directed Against the von Willebrand factor A3-Domain | | Descriptor: | 82D6A3 IgG, ACETIC ACID, GLYCEROL, ... | | Authors: | Staelens, S, Hadders, M.A, Vauterin, S, Platteau, C, Vanhoorelbeke, K, Huizinga, E.G, Deckmyn, H. | | Deposit date: | 2005-07-20 | | Release date: | 2005-12-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Paratope determination of the antithrombotic antibody 82D6A3 based on the crystal structure of its complex with the von Willebrand factor A3-domain
J.Biol.Chem., 281, 2006
|
|
1Y5C
 
 | |
2A1E
 
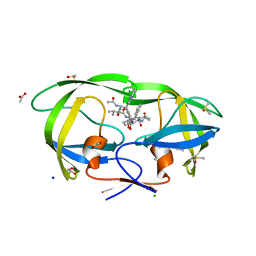 | | High resolution structure of HIV-1 PR with TS-126 | | Descriptor: | ACETATE ION, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Demitri, N, Geremia, S, Randaccio, L, Wuerges, J, Benedetti, F, Berti, F, Dinon, F, Campaner, P, Tell, G. | | Deposit date: | 2005-06-20 | | Release date: | 2006-02-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A potent HIV protease inhibitor identified in an epimeric mixture by high-resolution protein crystallography.
Chemmedchem, 1, 2006
|
|
1ZZH
 
 | | Structure of the fully oxidized di-heme cytochrome c peroxidase from R. capsulatus | | Descriptor: | CALCIUM ION, HEME C, ZINC ION, ... | | Authors: | De Smet, L, Savvides, S.N, Van Horen, E, Pettigrew, G, Van Beeumen, J.J. | | Deposit date: | 2005-06-14 | | Release date: | 2005-11-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and mutagenesis studies on the cytochrome c peroxidase from Rhodobacter capsulatus provide new insights into structure-function relationships of bacterial di-heme peroxidases
J.Biol.Chem., 281, 2006
|
|
2AFO
 
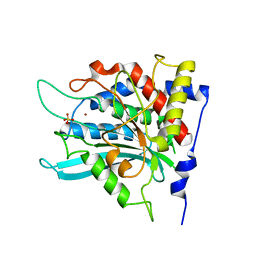 | | Crystal structure of human glutaminyl cyclase at pH 8.0 | | Descriptor: | Glutaminyl-peptide cyclotransferase, SULFATE ION, ZINC ION | | Authors: | Huang, K.F, Liu, Y.L, Cheng, W.J, Ko, T.P, Wang, A.H.J. | | Deposit date: | 2005-07-26 | | Release date: | 2005-08-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structures of human glutaminyl cyclase, an enzyme responsible for protein N-terminal pyroglutamate formation
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2AFW
 
 | | Crystal structure of human glutaminyl cyclase in complex with N-acetylhistamine | | Descriptor: | Glutaminyl-peptide cyclotransferase, N-[2-(1H-IMIDAZOL-4-YL)ETHYL]ACETAMIDE, SULFATE ION, ... | | Authors: | Huang, K.F, Liu, Y.L, Cheng, W.J, Ko, T.P, Wang, A.H.J. | | Deposit date: | 2005-07-26 | | Release date: | 2005-08-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal structures of human glutaminyl cyclase, an enzyme responsible for protein N-terminal pyroglutamate formation
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1UMX
 
 | | PHOTOSYNTHETIC REACTION CENTER MUTANT WITH ARG M267 REPLACED WITH LEU (CHAIN M, R267L) | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN B, FE (III) ION, ... | | Authors: | Fyfe, P.K, Isaacs, N.W, Cogdell, R.J, Jones, M.R. | | Deposit date: | 2003-09-02 | | Release date: | 2004-06-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Disruption of a specific molecular interaction with a bound lipid affects the thermal stability of the purple bacterial reaction centre.
Biochim.Biophys.Acta, 1608, 2004
|
|
2AFU
 
 | | Crystal structure of human glutaminyl cyclase in complex with glutamine t-butyl ester | | Descriptor: | Glutaminyl-peptide cyclotransferase, TERT-BUTYL D-ALPHA-GLUTAMINATE, ZINC ION | | Authors: | Huang, K.F, Liu, Y.L, Cheng, W.J, Ko, T.P, Wang, A.H.J. | | Deposit date: | 2005-07-26 | | Release date: | 2005-08-23 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structures of human glutaminyl cyclase, an enzyme responsible for protein N-terminal pyroglutamate formation
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
