8DQ2
 
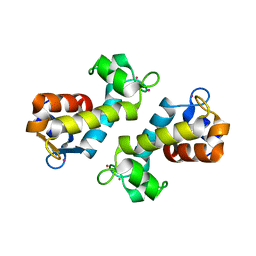 | | X-ray crystal structure of Hansschlegelia quercus lanmodulin (LanM) with lanthanum (III) bound at pH 7 | | Descriptor: | CITRIC ACID, EF-hand domain-containing protein, LANTHANUM (III) ION, ... | | Authors: | Jung, J.J, Lin, C.-Y, Boal, A.K. | | Deposit date: | 2022-07-18 | | Release date: | 2023-06-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Enhanced rare-earth separation with a metal-sensitive lanmodulin dimer.
Nature, 618, 2023
|
|
6OKT
 
 | |
8E2W
 
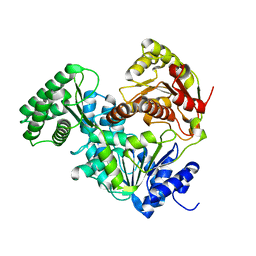 | | Structure of CRISPR-Associated DinG | | Descriptor: | CasDinG | | Authors: | Domgaard, H, Jackson, R.N. | | Deposit date: | 2022-08-16 | | Release date: | 2023-06-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | CasDinG is a 5'-3' dsDNA and RNA/DNA helicase with three accessory domains essential for type IV CRISPR immunity.
Nucleic Acids Res., 51, 2023
|
|
4WOD
 
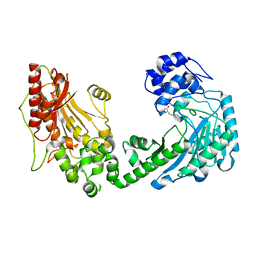 | | The duplicated taurocyamine kinase from Schistosoma mansoni complexed with arginine | | Descriptor: | ARGININE, Taurocyamine kinase | | Authors: | Merceron, R, Awama, A, Montserret, R, Marcillat, O, Gouet, P. | | Deposit date: | 2014-10-15 | | Release date: | 2015-04-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Substrate-free and -bound Crystal Structures of the Duplicated Taurocyamine Kinase from the Human Parasite Schistosoma mansoni.
J.Biol.Chem., 290, 2015
|
|
6O64
 
 | |
5KRS
 
 | | HIV-1 Integrase Catalytic Core Domain in Complex with an Allosteric Inhibitor, 3-(1H-pyrrol-1-yl)-2-thiophenecarboxylic acid | | Descriptor: | 3-pyrrol-1-ylthiophene-2-carboxylic acid, DIMETHYL SULFOXIDE, Integrase | | Authors: | Patel, D, Bauman, J.D, Arnold, E. | | Deposit date: | 2016-07-07 | | Release date: | 2016-09-28 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A New Class of Allosteric HIV-1 Integrase Inhibitors Identified by Crystallographic Fragment Screening of the Catalytic Core Domain.
J.Biol.Chem., 291, 2016
|
|
8DNG
 
 | |
8DNR
 
 | |
5KQV
 
 | | Insulin receptor ectodomain construct comprising domains L1,CR,L2, FnIII-1 and alphaCT peptide in complex with bovine insulin and FAB 83-14 (REVISED STRUCTURE) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Insulin, Insulin receptor,Insulin receptor, ... | | Authors: | Lawrence, M.C, Smith, B.J, Croll, T.I. | | Deposit date: | 2016-07-06 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | How insulin engages its primary binding site on the insulin receptor.
Nature, 493, 2013
|
|
8DO4
 
 | |
5KQA
 
 | | Crystal structure of buckwheat glutaredoxin-glutathione complex | | Descriptor: | GLUTATHIONE, Glutaredoxin-glutathione complex | | Authors: | Zhang, X, Wang, W, Zhao, Y, Wang, Z, Wang, H. | | Deposit date: | 2016-07-06 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural insights into the binding of buckwheat glutaredoxin with GSH and regulation of its catalytic activity
J. Inorg. Biochem., 173, 2017
|
|
5KVC
 
 | |
4X1W
 
 | | Crystal structure of unbound RHDVb P domain | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, VP1 | | Authors: | Leuthold, M.M, Hansman, G.S. | | Deposit date: | 2014-11-25 | | Release date: | 2015-01-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analysis of a rabbit hemorrhagic disease virus binding to histo-blood group antigens.
J.Virol., 89, 2015
|
|
5KLO
 
 | | Crystal structure of thioacyl intermediate in 2-aminomuconate 6-semialdehyde dehydrogenase N169A | | Descriptor: | (2Z,4E)-2-hydroxy-6-oxohexa-2,4-dienoic acid, 2-aminomuconate 6-semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Yang, Y, Davis, I, Ha, U, Wang, Y, Shin, I, Liu, A. | | Deposit date: | 2016-06-24 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A Pitcher-and-Catcher Mechanism Drives Endogenous Substrate Isomerization by a Dehydrogenase in Kynurenine Metabolism.
J.Biol.Chem., 291, 2016
|
|
5KRT
 
 | |
1OID
 
 | |
5L4M
 
 | | Crystal Structure of Human Transthyretin in Complex with 3,5,6-Trichloro-2-pyridinyloxyacetic acid (Triclopyr) | | Descriptor: | SODIUM ION, Transthyretin, Triclopyr | | Authors: | Grundstrom, C, Hall, M, Zhang, J, Olofsson, A, Andersson, P, Sauer-Eriksson, A.E. | | Deposit date: | 2016-05-25 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.581 Å) | | Cite: | Structure-Based Virtual Screening Protocol for in Silico Identification of Potential Thyroid Disrupting Chemicals Targeting Transthyretin.
Environ. Sci. Technol., 50, 2016
|
|
4WQ5
 
 | | YgjD(V85E)-YeaZ heterodimer in complex with ATP | | Descriptor: | ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, FE (III) ION, ... | | Authors: | Zhang, W. | | Deposit date: | 2014-10-21 | | Release date: | 2015-01-28 | | Last modified: | 2015-02-25 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | The ATP-mediated formation of the YgjD-YeaZ-YjeE complex is required for the biosynthesis of tRNA t6A in Escherichia coli.
Nucleic Acids Res., 43, 2015
|
|
4WQM
 
 | | Structure of the toluene 4-monooxygenase NADH oxidoreductase T4moF, K270S K271S variant | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Acheson, J.F, Fox, B.G. | | Deposit date: | 2014-10-22 | | Release date: | 2015-09-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure of T4moF, the Toluene 4-Monooxygenase Ferredoxin Oxidoreductase.
Biochemistry, 54, 2015
|
|
3LAD
 
 | |
5KLM
 
 | | Crystal structure of 2-hydroxymuconate-6-semialdehyde derived intermediate in NAD(+)-bound 2-aminomuconate 6-semialdehyde dehydrogenase N169D | | Descriptor: | 2-aminomuconate 6-semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION | | Authors: | Yang, Y, Davis, I, Ha, U, Wang, Y, Shin, I, Liu, A. | | Deposit date: | 2016-06-24 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | A Pitcher-and-Catcher Mechanism Drives Endogenous Substrate Isomerization by a Dehydrogenase in Kynurenine Metabolism.
J.Biol.Chem., 291, 2016
|
|
8DYD
 
 | | Crystal structure of human SDHA-SDHAF2-SDHAF4 assembly intermediate | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sharma, P, Maklashina, E, Cecchini, G, Iverson, T.M. | | Deposit date: | 2022-08-04 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Disordered-to-ordered transitions in assembly factors allow the complex II catalytic subunit to switch binding partners.
Nat Commun, 15, 2024
|
|
8DYE
 
 | | Crystal structure of human SDHA-SDHAF4 assembly intermediate | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Sharma, P, Maklashina, E, Cecchini, G, Iverson, T.M. | | Deposit date: | 2022-08-04 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Disordered-to-ordered transitions in assembly factors allow the complex II catalytic subunit to switch binding partners.
Nat Commun, 15, 2024
|
|
8ENC
 
 | | Helical reconstruction of the human cardiac actin-tropomyosin-myosin loop 4 7G mutant complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha cardiac muscle 1, ... | | Authors: | Doran, M.H, Lehman, W, Rynkiewicz, M.J. | | Deposit date: | 2022-09-29 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Myosin loop-4 is critical for optimal tropomyosin repositioning on actin during muscle activation and relaxation.
J.Gen.Physiol., 155, 2023
|
|
7KDV
 
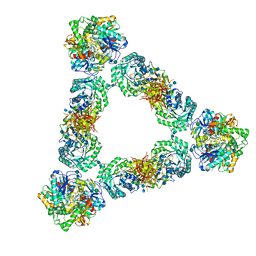 | | Murine core lysosomal multienzyme complex (LMC) composed of acid beta-galactosidase (GLB1) and protective protein cathepsin A (PPCA, CTSA) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-galactosidase, ... | | Authors: | Gorelik, A, Illes, K, Hasan, S.M.N, Nagar, B, Mazhab-Jafari, M.T. | | Deposit date: | 2020-10-09 | | Release date: | 2021-03-17 | | Last modified: | 2021-05-26 | | Method: | ELECTRON MICROSCOPY (4.59 Å) | | Cite: | Structure of the murine lysosomal multienzyme complex core.
Sci Adv, 7, 2021
|
|
