1WBN
 
 | | fragment based p38 inhibitors | | Descriptor: | MITOGEN-ACTIVATED PROTEIN KINASE 14, N-(3-TERT-BUTYL-1H-PYRAZOL-5-YL)-N'-{4-CHLORO-3-[(PYRIDIN-3-YLOXY)METHYL]PHENYL}UREA | | Authors: | Cleasby, A, Devine, L.A, Gill, A.L, Jhoti, H. | | Deposit date: | 2004-11-04 | | Release date: | 2005-11-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of Novel P38Alpha Map Kinase Inhibitors Using Fragment-Based Lead Generation.
J.Med.Chem., 48, 2005
|
|
5UDA
 
 | |
4IIT
 
 | |
5UGU
 
 | | Crystal structure of M. tuberculosis InhA inhibited by PT506 | | Descriptor: | 2-[4-[(4-cyclopropyl-1,2,3-triazol-1-yl)methyl]-2-oxidanyl-phenoxy]benzenecarbonitrile, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Eltschkner, S, Pschibul, A, Spagnuolo, L.A, Yu, W, Tonge, P.J, Kisker, C. | | Deposit date: | 2017-01-10 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Evaluating the Contribution of Transition-State Destabilization to Changes in the Residence Time of Triazole-Based InhA Inhibitors.
J. Am. Chem. Soc., 139, 2017
|
|
7Q5V
 
 | | HIF PROLYL HYDROXYLASE 2 (PHD2/EGLN1) IN COMPLEX WITH N-OXALYLGLYCINE (NOG) AND HIF-2 ALPHA CODD (523-542) | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Egl nine homolog 1, ... | | Authors: | Figg Jr, W.D, McDonough, M.A, Chowdhury, R, Nakashima, Y, Schofield, C.J. | | Deposit date: | 2021-11-04 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Structural basis for binding of the renal carcinoma target hypoxia-inducible factor 2 alpha to prolyl hydroxylase domain 2.
Proteins, 91, 2023
|
|
4EZH
 
 | |
5UGT
 
 | | Crystal structure of M. tuberculosis InhA inhibited by PT504 | | Descriptor: | 2-(2-chloranylphenoxy)-5-[(4-cyclopropyl-1,2,3-triazol-1-yl)methyl]phenol, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Eltschkner, S, Pschibul, A, Spagnuolo, L.A, Yu, W, Tonge, P.J, Kisker, C. | | Deposit date: | 2017-01-10 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Evaluating the Contribution of Transition-State Destabilization to Changes in the Residence Time of Triazole-Based InhA Inhibitors.
J. Am. Chem. Soc., 139, 2017
|
|
4ICT
 
 | | Substrate and reaction specificity of Mycobacterium tuberculosis cytochrome P450 CYP121 | | Descriptor: | (3S,6S)-3-benzyl-6-(4-hydroxybenzyl)piperazine-2,5-dione, Cytochrome P450 121, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Fonvielle, M, Le Du, M.-H, Lequin, O, Lecoq, A, Jacquet, M, Thai, R, Dubois, S, Grach, G, Gondry, M, Belin, P. | | Deposit date: | 2012-12-11 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Substrate and Reaction Specificity of Mycobacterium tuberculosis Cytochrome P450 CYP121: INSIGHTS FROM BIOCHEMICAL STUDIES AND CRYSTAL STRUCTURES.
J.Biol.Chem., 288, 2013
|
|
7Q6V
 
 | | Crystal structure of the bromodomain of ATAD2 with triazolopyridine (cpd 14) | | Descriptor: | (1R,9S)-13-[(8-azanyl-3-methyl-[1,2,4]triazolo[4,3-a]pyridin-6-yl)carbonyl]-11,13-diazatricyclo[7.3.1.0^{2,7}]trideca-2,4,6-trien-10-one, ATPase family AAA domain-containing protein 2, SULFATE ION | | Authors: | Patel, S.J, Winter-Holt, J.J. | | Deposit date: | 2021-11-09 | | Release date: | 2022-01-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Discovery of a Potent and Selective ATAD2 Bromodomain Inhibitor with Antiproliferative Activity in Breast Cancer Models.
J.Med.Chem., 65, 2022
|
|
5UNY
 
 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with (RS)-3-[(2-amino-4-methylquinolin-7-yl)methoxy]-5-(2-(methylamino)propyl)benzonitrile | | Descriptor: | 3-[(2-amino-4-methylquinolin-7-yl)methoxy]-5-[(2R)-2-(methylamino)propyl]benzonitrile, 3-[(2-amino-4-methylquinolin-7-yl)methoxy]-5-[(2S)-2-(methylamino)propyl]benzonitrile, 5,6,7,8-TETRAHYDROBIOPTERIN, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2017-01-31 | | Release date: | 2017-05-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Nitrile in the Hole: Discovery of a Small Auxiliary Pocket in Neuronal Nitric Oxide Synthase Leading to the Development of Potent and Selective 2-Aminoquinoline Inhibitors.
J. Med. Chem., 60, 2017
|
|
1WBO
 
 | | fragment based p38 inhibitors | | Descriptor: | 2-CHLOROPHENOL, MITOGEN-ACTIVATED PROTEIN KINASE 14 | | Authors: | Cleasby, A, Devine, L, Gill, A, Jhoti, H. | | Deposit date: | 2004-11-04 | | Release date: | 2005-01-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Fragment-Based Lead Discovery Using X-Ray Crystallography
J.Med.Chem., 48, 2005
|
|
1WBW
 
 | | Identification of novel p38 alpha MAP Kinase inhibitors using fragment-based lead generation. | | Descriptor: | 3-(1-NAPHTHYLMETHOXY)PYRIDIN-2-AMINE, MITOGEN-ACTIVATED PROTEIN KINASE 14 | | Authors: | Tickle, J, Cleasby, A, Devine, L.A, Jhoti, H. | | Deposit date: | 2004-11-05 | | Release date: | 2005-11-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Identification of Novel P38Alpha Map Kinase Inhibitors Using Fragment-Based Lead Generation
J.Med.Chem., 48, 2005
|
|
4IDZ
 
 | | Crystal structure of the human fat mass and obesity associated protein (FTO) in complex with N-oxalylglycine (NOG) | | Descriptor: | Alpha-ketoglutarate-dependent dioxygenase FTO, GLYCEROL, N-OXALYLGLYCINE, ... | | Authors: | Aik, W.S, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2012-12-13 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structural basis for inhibition of the fat mass and obesity associated protein (FTO)
J.Med.Chem., 56, 2013
|
|
1WBV
 
 | | Identification of novel p38 alpha MAP Kinase inhibitors using fragment-based lead generation. | | Descriptor: | 3-FLUORO-N-1H-INDOL-5-YL-5-MORPHOLIN-4-YLBENZAMIDE, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Tickle, J, Cleasby, A, Devine, L.A, Jhoti, H. | | Deposit date: | 2004-11-05 | | Release date: | 2005-11-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of Novel P38Alpha Map Kinase Inhibitors Using Fragment-Based Lead Generation.
J.Med.Chem., 48, 2005
|
|
5UNR
 
 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 3-[(2-aminoquinolin-7-yl)methoxy]-5-((methylamino)methyl)benzonitrile | | Descriptor: | 3-[(2-aminoquinolin-7-yl)methoxy]-5-[(methylamino)methyl]benzonitrile, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2017-01-31 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Nitrile in the Hole: Discovery of a Small Auxiliary Pocket in Neuronal Nitric Oxide Synthase Leading to the Development of Potent and Selective 2-Aminoquinoline Inhibitors.
J. Med. Chem., 60, 2017
|
|
5UIT
 
 | | Crystal structure of IRAK4 in complex with compound 14 | | Descriptor: | 1-{[(2S)-5-oxopyrrolidin-2-yl]methoxy}-7-[(propan-2-yl)oxy]isoquinoline-6-carboxamide, Interleukin-1 receptor-associated kinase 4 | | Authors: | Han, S, Chang, J.S. | | Deposit date: | 2017-01-14 | | Release date: | 2017-05-24 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Discovery of Clinical Candidate 1-{[(2S,3S,4S)-3-Ethyl-4-fluoro-5-oxopyrrolidin-2-yl]methoxy}-7-methoxyisoquinoline-6-carboxamide (PF-06650833), a Potent, Selective Inhibitor of Interleukin-1 Receptor Associated Kinase 4 (IRAK4), by Fragment-Based Drug Design.
J. Med. Chem., 60, 2017
|
|
4J2W
 
 | | Crystal Structure of kynurenine 3-monooxygenase (KMO-396Prot-Se) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Kynurenine 3-monooxygenase | | Authors: | Amaral, M, Levy, C, Heyes, D.J, Lafite, P, Outeiro, T.F, Giorgini, F, Leys, D, Scrutton, N.S. | | Deposit date: | 2013-02-05 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of kynurenine 3-monooxygenase inhibition.
Nature, 496, 2013
|
|
4FSF
 
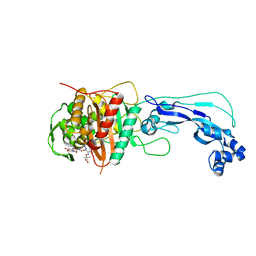 | | Crystal structure of Pseudomonas aeruginosa PBP3 complexed with compound 14 | | Descriptor: | (4R,5S,8Z)-8-(2-amino-1,3-thiazol-4-yl)-1-[3-(1,5-dihydroxy-4-oxo-1,4-dihydropyridin-2-yl)-1,2-oxazol-5-yl]-5-formyl-11,11-dimethyl-1,7-dioxo-4-(sulfoamino)-10-oxa-2,6,9-triazadodec-8-en-12-oic acid, Penicillin-binding protein 3 | | Authors: | Han, S. | | Deposit date: | 2012-06-27 | | Release date: | 2012-10-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Novel monobactams utilizing a siderophore uptake mechanism for the treatment of gram-negative infections.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
5UNT
 
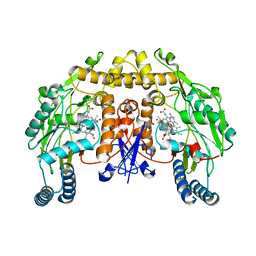 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 4-Methyl-7-[(3-((methylamino)methyl)phenoxy)methyl]quinolin-2-amine | | Descriptor: | 4-methyl-7-({3-[(methylamino)methyl]phenoxy}methyl)quinolin-2-amine, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2017-01-31 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Nitrile in the Hole: Discovery of a Small Auxiliary Pocket in Neuronal Nitric Oxide Synthase Leading to the Development of Potent and Selective 2-Aminoquinoline Inhibitors.
J. Med. Chem., 60, 2017
|
|
4G32
 
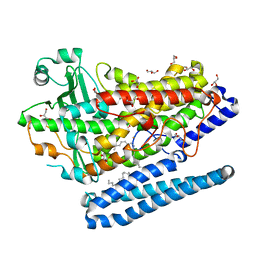 | | Crystal Structure of a Phospholipid-Lipoxygenase Complex from Pseudomonas aeruginosa at 1.75A (P21212) | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradec-5-enoyloxy)propyl (11Z)-octadec-11-enoate, 15S-LIPOXYGENASE, FE (II) ION, ... | | Authors: | Carpena, X, Garreta, A, Val-Moraes, S.P, Garcia-Fernandez, Q, Fita, I. | | Deposit date: | 2012-07-13 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and interaction with phospholipids of a prokaryotic lipoxygenase from Pseudomonas aeruginosa.
Faseb J., 27, 2013
|
|
4G0D
 
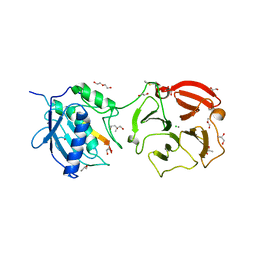 | | Human collagenase 3 (MMP-13) full form with peptides from pro-domain | | Descriptor: | CALCIUM ION, CHLORIDE ION, Collagenase 3, ... | | Authors: | Stura, E.A, Vera, L, Visse, R, Nagase, H, Dive, V. | | Deposit date: | 2012-07-09 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Crystal structure of full-length human collagenase 3 (MMP-13) with peptides in the active site defines exosites in the catalytic domain.
Faseb J., 27, 2013
|
|
4G78
 
 | |
5VUP
 
 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 7-(((3-(5-Fluoropyridin-3-yl)propyl)amino)methyl)quinolin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-({[3-(5-fluoropyridin-3-yl)propyl]amino}methyl)quinolin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2017-05-19 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Hydrophilic, Potent, and Selective 7-Substituted 2-Aminoquinolines as Improved Human Neuronal Nitric Oxide Synthase Inhibitors.
J. Med. Chem., 60, 2017
|
|
7OO5
 
 | |
4GAZ
 
 | | Crystal Structure of a Jumonji Domain-containing Protein JMJD5 | | Descriptor: | Lysine-specific demethylase 8, N-OXALYLGLYCINE, NICKEL (II) ION | | Authors: | Wang, H, Zhou, X, Zhang, X, Tao, Y, Chen, N, Zang, J. | | Deposit date: | 2012-07-26 | | Release date: | 2013-08-14 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Crystal Structure of a Jumonji Domain-containing Protein JMJD5
To be Published
|
|
