8IWS
 
 | | hSPCA1 in the CaE2P state | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, CALCIUM ION, Calcium-transporting ATPase type 2C member 1, ... | | Authors: | Liu, Z.M, Wu, M.Q, Wu, C. | | Deposit date: | 2023-03-31 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Structure and transport mechanism of the human calcium pump SPCA1.
Cell Res., 33, 2023
|
|
8IWR
 
 | | hSPCA1 in the CaE1-ATP state | | Descriptor: | CALCIUM ION, Calcium-transporting ATPase type 2C member 1, MAGNESIUM ION, ... | | Authors: | Liu, Z.M, Wu, M.Q, Wu, C. | | Deposit date: | 2023-03-31 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Structure and transport mechanism of the human calcium pump SPCA1.
Cell Res., 33, 2023
|
|
8IWP
 
 | | hSPCA1 in the CaE1 state | | Descriptor: | CALCIUM ION, Calcium-transporting ATPase type 2C member 1 | | Authors: | Liu, Z.M, Wu, M.Q, Wu, C. | | Deposit date: | 2023-03-30 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Structure and transport mechanism of the human calcium pump SPCA1.
Cell Res., 33, 2023
|
|
8DFM
 
 | | Ectodomain of full-length wild-type KIT-SCF dimers | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform 2 of Mast/stem cell growth factor receptor Kit, ... | | Authors: | Krimmer, S.G, Bertoletti, N, Mi, W, Schlessinger, J. | | Deposit date: | 2022-06-22 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Cryo-EM analyses of KIT and oncogenic mutants reveal structural oncogenic plasticity and a target for therapeutic intervention.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8DCN
 
 | |
8DFQ
 
 | | Ectodomain of full-length KIT(T417I,delta418-419)-SCF dimers | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform 2 of Mast/stem cell growth factor receptor Kit, ... | | Authors: | Krimmer, S.G, Bertoletti, N, Mi, W, Schlessinger, J. | | Deposit date: | 2022-06-22 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Cryo-EM analyses of KIT and oncogenic mutants reveal structural oncogenic plasticity and a target for therapeutic intervention.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8DCM
 
 | |
8JNJ
 
 | | Structure of R932A/K1147A/H1148A mutant AE2 | | Descriptor: | Anion exchange protein 2 | | Authors: | Yin, Y.X, Ding, D. | | Deposit date: | 2023-06-06 | | Release date: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural and functional insights into the lipid regulation of human anion exchanger 2.
Nat Commun, 15, 2024
|
|
8JNI
 
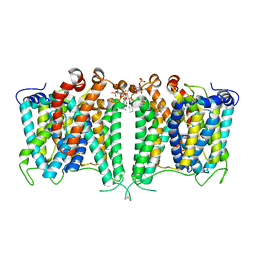 | | Structure of AE2 in complex with PIP2 | | Descriptor: | Anion exchange protein 2, CHLORIDE ION, [(2R)-1-octadecanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phospho ryl]oxy-propan-2-yl] (8Z)-icosa-5,8,11,14-tetraenoate | | Authors: | Yin, Y.X, Ding, D. | | Deposit date: | 2023-06-06 | | Release date: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and functional insights into the lipid regulation of human anion exchanger 2.
Nat Commun, 15, 2024
|
|
8ITV
 
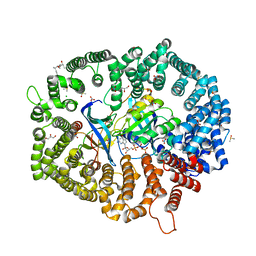 | | KL2.1 in complex with CRM1-Ran-RanBP1 | | Descriptor: | (3~{S},5~{R},6~{E},8~{Z},10~{R},12~{E},14~{E},16~{S})-3,16-bis(azanyl)-8,10,12-trimethyl-16-[(2~{S},4~{R},5~{S},6~{S})-5-methyl-4-oxidanyl-6-[(~{E})-prop-1-enyl]oxan-2-yl]-5-oxidanyl-hexadeca-6,8,12,14-tetraenoic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Sun, Q, Jian, L. | | Deposit date: | 2023-03-23 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | KL2.1 in complex with CRM1-Ran-RanBP1
To Be Published
|
|
7UTH
 
 | |
7UT4
 
 | |
8D4F
 
 | | beta-Arf1 mediated dimeric assembly of AP-1, Arf1, Nef complex within lattice on MHC-I lipopeptide incorporated wide(r) membrane tubes | | Descriptor: | ADP-ribosylation factor 1, AP-1 complex subunit beta-1, AP-1 complex subunit gamma-1, ... | | Authors: | Hooy, R.M, Hurley, J.H. | | Deposit date: | 2022-06-01 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (9.8 Å) | | Cite: | Self-assembly and structure of a clathrin-independent AP-1:Arf1 tubular membrane coat.
Sci Adv, 8, 2022
|
|
8D4E
 
 | | Asymmetric unit of AP-1, Arf1, Nef lattice on MHC-I lipopeptide incorporated wide(r) membrane tubes | | Descriptor: | ADP-ribosylation factor 1, AP-1 complex subunit beta-1, AP-1 complex subunit gamma-1, ... | | Authors: | Hooy, R.M, Hurley, J.H. | | Deposit date: | 2022-06-01 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (9.2 Å) | | Cite: | Self-assembly and structure of a clathrin-independent AP-1:Arf1 tubular membrane coat.
Sci Adv, 8, 2022
|
|
8D4G
 
 | | gamma-Arf1 mediated dimeric assembly of AP-1, Arf1, Nef complex within lattice on MHC-I lipopeptide incorporated wide(r) membrane tubes | | Descriptor: | ADP-ribosylation factor 1, AP-1 complex subunit beta-1, AP-1 complex subunit gamma-1, ... | | Authors: | Hooy, R.M, Hurley, J.H. | | Deposit date: | 2022-06-01 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (11.6 Å) | | Cite: | Self-assembly and structure of a clathrin-independent AP-1:Arf1 tubular membrane coat.
Sci Adv, 8, 2022
|
|
8D4D
 
 | | gamma-Arf1 mediated dimeric assembly of AP-1, Arf1, Nef complex within lattice on MHC-I lipopeptide incorporated narrow membrane tubes | | Descriptor: | ADP-ribosylation factor 1, AP-1 complex subunit beta-1, AP-1 complex subunit gamma-1, ... | | Authors: | Hooy, R.M, Hurley, J.H. | | Deposit date: | 2022-06-01 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (9.6 Å) | | Cite: | Self-assembly and structure of a clathrin-independent AP-1:Arf1 tubular membrane coat.
Sci Adv, 8, 2022
|
|
8D4C
 
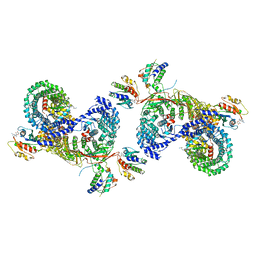 | | beta-Arf1 mediated dimeric assembly of AP-1, Arf1, Nef complex within lattice on MHC-I lipopeptide incorporated narrow membrane tubes | | Descriptor: | ADP-ribosylation factor 1, AP-1 complex subunit beta-1, AP-1 complex subunit gamma-1, ... | | Authors: | Hooy, R.M, Hurley, J.H. | | Deposit date: | 2022-06-01 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (9.3 Å) | | Cite: | Self-assembly and structure of a clathrin-independent AP-1:Arf1 tubular membrane coat.
Sci Adv, 8, 2022
|
|
8D9W
 
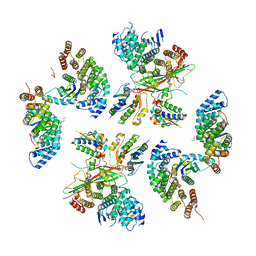 | | beta-Arf1 homodimeric interface within AP-1, Arf1, Nef, MHC-I lattice on narrow tubes | | Descriptor: | ADP-ribosylation factor 1, AP-1 complex subunit beta-1, AP-1 complex subunit gamma-1, ... | | Authors: | Hooy, R.H, Hurley, J.H. | | Deposit date: | 2022-06-11 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (9.3 Å) | | Cite: | Self-assembly and structure of a clathrin-independent AP-1:Arf1 tubular membrane coat.
Sci Adv, 8, 2022
|
|
2D88
 
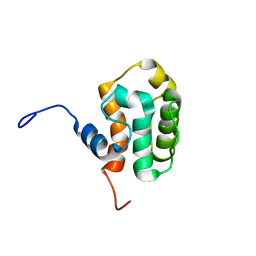 | | Solution structure of the CH domain from human MICAL-3 protein | | Descriptor: | Protein MICAL-3 | | Authors: | Tomizawa, T, Kigawa, T, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-02 | | Release date: | 2006-06-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the CH domain from human MICAL-3 protein
To be Published
|
|
8J8Y
 
 | |
7UHE
 
 | |
8DVR
 
 | | Cryo-EM structure of RIG-I bound to the end of p3SLR30 (+AMPPNP) | | Descriptor: | Antiviral innate immune response receptor RIG-I, GUANOSINE-5'-TRIPHOSPHATE, ZINC ION, ... | | Authors: | Wang, W, Pyle, A.M. | | Deposit date: | 2022-07-29 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The RIG-I receptor adopts two different conformations for distinguishing host from viral RNA ligands.
Mol.Cell, 82, 2022
|
|
8DVS
 
 | | Cryo-EM structure of RIG-I bound to the end of OHSLR30 (+ATP) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Antiviral innate immune response receptor RIG-I, MAGNESIUM ION, ... | | Authors: | Wang, W, Pyle, A.M. | | Deposit date: | 2022-07-29 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The RIG-I receptor adopts two different conformations for distinguishing host from viral RNA ligands.
Mol.Cell, 82, 2022
|
|
7UR2
 
 | |
8DVU
 
 | |
