1LUG
 
 | | Full Matrix Error Analysis of Carbonic Anhydrase | | Descriptor: | (4-SULFAMOYL-PHENYL)-THIOCARBAMIC ACID O-(2-THIOPHEN-3-YL-ETHYL) ESTER, Carbonic Anhydrase II, GLYCEROL, ... | | Authors: | Merritt, E.A, Le Trong, I, Behnke, C.A. | | Deposit date: | 2002-05-22 | | Release date: | 2003-09-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Atomic resolution studies of carbonic anhydrase II.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
4QRS
 
 | | Crystal Structure of HLA B*0801 in complex with ELK_IYM, ELKRKMIYM | | Descriptor: | ACETATE ION, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Gras, S, Twist, K.-A, Rossjohn, J. | | Deposit date: | 2014-07-02 | | Release date: | 2014-12-10 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular imprint of exposure to naturally occurring genetic variants of human cytomegalovirus on the T cell repertoire.
Sci Rep, 4, 2014
|
|
4LQR
 
 | | Structure of CBM32-3 from a family 31 glycoside hydrolase from Clostridium perfringens | | Descriptor: | CALCIUM ION, Glycosyl hydrolase, family 31/fibronectin type III domain protein | | Authors: | Grondin, J.M, Furness, H.S, Duan, D, Spencer, C.A, Allingham, J.S, Smith, S.P. | | Deposit date: | 2013-07-19 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Diverse modes of galacto-specific carbohydrate recognition by a family 31 glycoside hydrolase from Clostridium perfringens.
Plos One, 12, 2017
|
|
3AQU
 
 | | Crystal structure of a class V chitinase from Arabidopsis thaliana | | Descriptor: | At4g19810, CITRATE ANION | | Authors: | Numata, T, Ohnuma, T, Osawa, T, Fukamizo, T. | | Deposit date: | 2010-11-19 | | Release date: | 2011-07-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | A class V chitinase from Arabidopsis thaliana: gene responses, enzymatic properties, and crystallographic analysis
Planta, 234, 2011
|
|
1LY3
 
 | | ANALYSIS OF QUINAZOLINE AND PYRIDOPYRIMIDINE N9-C10 REVERSED BRIDGE ANTIFOLATES IN COMPLEX WITH NADP+ AND PNEUMOCYSTIS CARINII DIHYDROFOLATE REDUCTASE | | Descriptor: | 2,4-DIAMINO-6-[N-(2',5'-DIMETHOXYBENZYL)-N-METHYLAMINO]QUINAZOLINE, DIHYDROFOLATE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Galitsky, N, Luft, J.R, Pangborn, W, Queener, S.F, Gangjee, A. | | Deposit date: | 2002-06-06 | | Release date: | 2002-08-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Analysis of quinazoline and pyrido[2,3-d]pyrimidine N9-C10 reversed-bridge antifolates in complex with NADP+ and Pneumocystis carinii dihydrofolate reductase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
4MV6
 
 | | Crystal Structure of Biotin Carboxylase from Haemophilus influenzae in Complex with Phosphonoacetamide | | Descriptor: | 1,2-ETHANEDIOL, Biotin carboxylase, PHOSPHONOACETAMIDE | | Authors: | Broussard, T.C, Pakhomova, S, Neau, D.B, Champion, T.S, Bonnot, R.J, Waldrop, G.L. | | Deposit date: | 2013-09-23 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural Analysis of Substrate, Reaction Intermediate, and Product Binding in Haemophilus influenzae Biotin Carboxylase.
Biochemistry, 54, 2015
|
|
4MV9
 
 | | Crystal Structure of Biotin Carboxylase from Haemophilus influenzae in Complex with Bicarbonate | | Descriptor: | 1,2-ETHANEDIOL, BICARBONATE ION, Biotin carboxylase | | Authors: | Broussard, T.C, Pakhomova, S, Neau, D.B, Champion, T.S, Bonnot, R.J, Waldrop, G.L. | | Deposit date: | 2013-09-23 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural Analysis of Substrate, Reaction Intermediate, and Product Binding in Haemophilus influenzae Biotin Carboxylase.
Biochemistry, 54, 2015
|
|
4MWD
 
 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor 1-{3-[(3,5-dichlorobenzyl)amino]propyl}-3-thiophen-3-ylurea (Chem 1433) and ATP analog AMPPCP | | Descriptor: | 1-{3-[(3,5-dichlorobenzyl)amino]propyl}-3-thiophen-3-ylurea, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Koh, C.Y, Kim, J.E, Wetzel, A.B, de van der Schueren, W.J, Shibata, S, Liu, J, Zhang, Z, Fan, E, Verlinde, C.L.M.J, Hol, W.G.J. | | Deposit date: | 2013-09-24 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.253 Å) | | Cite: | Structures of Trypanosoma brucei Methionyl-tRNA Synthetase with Urea-Based Inhibitors Provide Guidance for Drug Design against Sleeping Sickness.
Plos Negl Trop Dis, 8, 2014
|
|
1J6U
 
 | |
4NQX
 
 | |
3AAR
 
 | | Crystal structure of Lp1NTPDase from Legionella pneumophila in complex with AMPPNP | | Descriptor: | Ectonucleoside triphosphate diphosphohydrolase I, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Ge, H, Vivian, J.P, Beddoe, T, Rossjohn, J. | | Deposit date: | 2009-11-24 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of a Legionella pneumophila Ecto -Triphosphate Diphosphohydrolase, A Structural and Functional Homolog of the Eukaryotic NTPDases
Structure, 18, 2010
|
|
4NWK
 
 | | Crystal structure of hepatis c virus protease (ns3) complexed with bms-605339 aka n-(tert-butoxycarbonyl)-3-me thyl-l-valyl-(4r)-n-((1r,2s)-1-((cyclopropylsulfonyl)carba moyl)-2-vinylcyclopropyl)-4-((6-methoxy-1-isoquinolinyl)ox y)-l-prolinamide | | Descriptor: | GLYCEROL, HCV NS3 1a Protease, N-(tert-butoxycarbonyl)-3-methyl-L-valyl-(4R)-N-{(1R,2S)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethenylcyclopropyl}-4-[(6-methoxyisoquinolin-1-yl)oxy]-L-prolinamide, ... | | Authors: | Muckelbauer, J.K, Klei, H.E. | | Deposit date: | 2013-12-06 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Discovery and Early Clinical Evaluation of BMS-605339, a Potent and Orally Efficacious Tripeptidic Acylsulfonamide NS3 Protease Inhibitor for the Treatment of Hepatitis C Virus Infection.
J.Med.Chem., 57, 2014
|
|
4NYM
 
 | | Approach for Targeting Ras with Small Molecules that Activate SOS-Mediated Nucleotide Exchange | | Descriptor: | GTPase HRas, MAGNESIUM ION, N-[1-(1H-indol-3-ylmethyl)piperidin-4-yl]-L-tryptophanamide, ... | | Authors: | Burns, M.C, Sun, Q, Phan, J, Fesik, S.W. | | Deposit date: | 2013-12-10 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5529 Å) | | Cite: | Approach for targeting Ras with small molecules that activate SOS-mediated nucleotide exchange.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3A3X
 
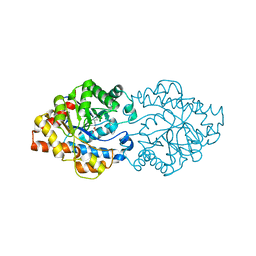 | | Structure of OpdA mutant (G60A/A80V/R118Q/K185R/Q206P/D208G/I260T/G273S) | | Descriptor: | COBALT (II) ION, Phosphotriesterase | | Authors: | Ollis, D.L, Tawfik, D.S, Schenk, G, Jackson, C.J, Foo, J.L, Tokuriki, N, Afriat, L, Carr, P.D, Kim, H.K. | | Deposit date: | 2009-06-23 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Conformational sampling, catalysis, and evolution of the bacterial phosphotriesterase
Proc.Natl.Acad.Sci.USA, 2009
|
|
1ZKG
 
 | |
3A4J
 
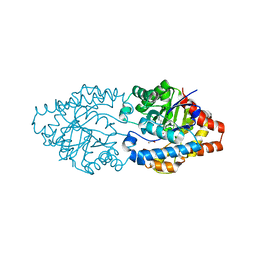 | | arPTE (K185R/D208G/N265D/T274N) | | Descriptor: | 1,2-ETHANEDIOL, COBALT (II) ION, Phosphotriesterase | | Authors: | Foo, J.L, Jackson, C.J, Carr, P.D, Ollis, D.L. | | Deposit date: | 2009-07-07 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Conformational sampling, catalysis, and evolution of the bacterial phosphotriesterase
Proc.Natl.Acad.Sci.USA, 2009
|
|
1MAJ
 
 | | SOLUTION STRUCTURE OF AN ISOLATED ANTIBODY VL DOMAIN | | Descriptor: | IGG2A-KAPPA 26-10 FV (LIGHT CHAIN) | | Authors: | Constantine, K.L, Friedrichs, M.S, Metzler, W.J, Wittekind, M, Hensley, P, Mueller, L. | | Deposit date: | 1993-09-16 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an isolated antibody VL domain.
J.Mol.Biol., 236, 1994
|
|
2ACY
 
 | |
4NCG
 
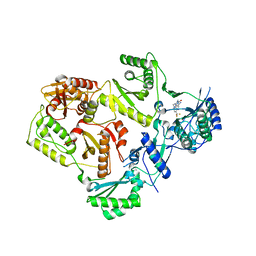 | | Discovery of Doravirine, an orally bioavailable non-nucleoside reverse transcriptase inhibitor potent against a wide range of resistant mutant HIV viruses | | Descriptor: | 3-chloro-5-({1-[(4-methyl-5-oxo-4,5-dihydro-1H-1,2,4-triazol-3-yl)methyl]-2-oxo-4-(trifluoromethyl)-1,2-dihydropyridin-3-yl}oxy)benzonitrile, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Yan, Y. | | Deposit date: | 2013-10-24 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Discovery of MK-1439, an orally bioavailable non-nucleoside reverse transcriptase inhibitor potent against a wide range of resistant mutant HIV viruses.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4K0O
 
 | | F17b-G lectin domain with bound GlcNAc(beta1-3)Gal | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-methyl beta-D-galactopyranoside, F17b-G fimbrial adhesin, NICKEL (II) ION, ... | | Authors: | Buts, L, Loris, R, Bouckaert, J, Moonens, K. | | Deposit date: | 2013-04-04 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Sampling of Glycan Interaction Profiles Reveals Mucosal Receptors for Fimbrial Adhesins of Enterotoxigenic Escherichia coli
Biology (Basel), 2, 2013
|
|
4K1F
 
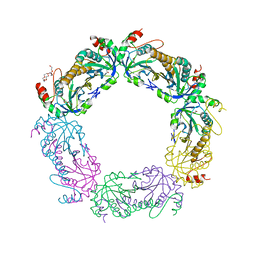 | | Crystal structure of reduced tryparedoxin peroxidase from leishmania major at 2.34 A resolution | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ilari, A, Fiorillo, A, Di Chiaro, F. | | Deposit date: | 2013-04-05 | | Release date: | 2014-04-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structure-based discovery of the first non-covalent inhibitors of Leishmania major tryparedoxin peroxidase by high throughput docking
Sci Rep, 5, 2015
|
|
4KA9
 
 | |
1MAK
 
 | | SOLUTION STRUCTURE OF AN ISOLATED ANTIBODY VL DOMAIN | | Descriptor: | IGG2A-KAPPA 26-10 FV (LIGHT CHAIN) | | Authors: | Constantine, K.L, Friedrichs, M.S, Metzler, W.J, Wittekind, M, Hensley, P, Mueller, L. | | Deposit date: | 1993-09-16 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an isolated antibody VL domain.
J.Mol.Biol., 236, 1994
|
|
4NYJ
 
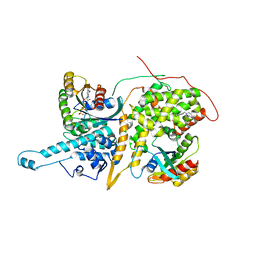 | | Approach for Targeting Ras with Small Molecules that Activate SOS-Mediated Nucleotide Exchange | | Descriptor: | GTPase HRas, MAGNESIUM ION, N-[1-(1H-indol-3-ylmethyl)piperidin-4-yl]glycinamide, ... | | Authors: | Burns, M.C, Sun, Q, Phan, J, Fesik, S.W. | | Deposit date: | 2013-12-10 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8522 Å) | | Cite: | Approach for targeting Ras with small molecules that activate SOS-mediated nucleotide exchange.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4NYI
 
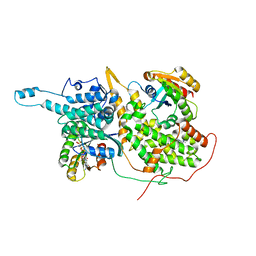 | | Approach for Targeting Ras with Small Molecules that Activate SOS-Mediated Nucleotide Exchange | | Descriptor: | GTPase HRas, MAGNESIUM ION, N-{1-[(5-methyl-1H-indol-3-yl)methyl]piperidin-4-yl}-L-tryptophanamide, ... | | Authors: | Burns, M.C, Sun, Q, Phan, J, Fesik, S.W. | | Deposit date: | 2013-12-10 | | Release date: | 2014-03-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9612 Å) | | Cite: | Approach for targeting Ras with small molecules that activate SOS-mediated nucleotide exchange.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
