8Y55
 
 | | Crystal Structure of NAMPT-PF403 | | Descriptor: | (13~{a}~{S})-6,7-dimethoxy-9,11,12,13,13~{a},14-hexahydrophenanthro[9,10-f]indolizin-3-ol, DI(HYDROXYETHYL)ETHER, Nicotinamide phosphoribosyltransferase | | Authors: | Yu, S.S, Liu, Y.B, Li, Y. | | Deposit date: | 2024-01-31 | | Release date: | 2025-02-19 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Thermal proteome profiling (TPP) reveals NAMPT as the anti-glioma target of phenanthroindolizidine alkaloid PF403.
Acta Pharm Sin B, 15, 2025
|
|
6H3H
 
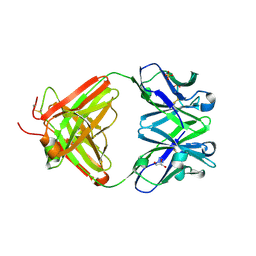 | | Fab fragment of antibody against fullerene C60 | | Descriptor: | Anti-fullerene antibody Fab fragment Heavy chain, Anti-fullerene antibody Fab fragment Light chain, GLYCEROL, ... | | Authors: | Osipov, E.M, Tikhonova, T.V. | | Deposit date: | 2018-07-18 | | Release date: | 2019-05-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure of the Anti-C60 Fullerene Antibody Fab Fragment: Structural Determinants of Fullerene Binding.
Acta Naturae, 11, 2019
|
|
2W3M
 
 | |
7DCN
 
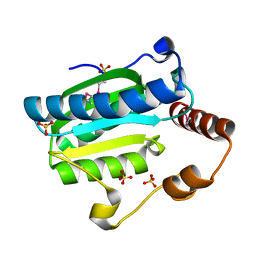 | | Apo-citrate lyase phosphoribosyl-dephospho-CoA transferase | | Descriptor: | Probable apo-citrate lyase phosphoribosyl-dephospho-CoA transferase, SULFATE ION, ZINC ION | | Authors: | Xu, H, Wang, B, Su, X.D. | | Deposit date: | 2020-10-26 | | Release date: | 2021-11-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | Co-evolution-based prediction of metal-binding sites in proteomes by machine learning.
Nat.Chem.Biol., 19, 2023
|
|
5J7I
 
 | |
6TOR
 
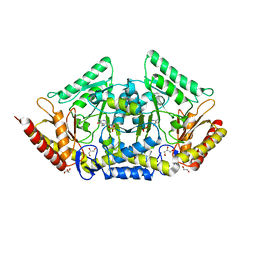 | | human O-phosphoethanolamine phospho-lyase | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Ethanolamine-phosphate phospho-lyase, GLYCEROL | | Authors: | Vettraino, C, Donini, S, Parisini, E. | | Deposit date: | 2019-12-11 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural characterization of human O-phosphoethanolamine phospho-lyase.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
7DCM
 
 | | Crystal structure of CITX | | Descriptor: | Probable apo-citrate lyase phosphoribosyl-dephospho-CoA transferase, ZINC ION | | Authors: | Xu, H, Wang, B, Su, X.D. | | Deposit date: | 2020-10-26 | | Release date: | 2021-11-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | Co-evolution-based prediction of metal-binding sites in proteomes by machine learning.
Nat.Chem.Biol., 19, 2023
|
|
6TQQ
 
 | |
6TQP
 
 | | Structural insight into tanapoxvirus mediated inhibition of apoptosis | | Descriptor: | 16L protein, Bcl-2-binding component 3, isoforms 1/2, ... | | Authors: | Suraweera, C.D, Hinds, M.G, Kvansakul, M. | | Deposit date: | 2019-12-17 | | Release date: | 2020-06-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.84940946 Å) | | Cite: | Structural insight into tanapoxvirus-mediated inhibition of apoptosis.
Febs J., 287, 2020
|
|
6TRR
 
 | |
6T76
 
 | | PII-like protein CutA from Nostoc sp. PCC 7120 in apo form | | Descriptor: | Periplasmic divalent cation tolerance protein, SULFATE ION | | Authors: | Selim, K.A, Albrecht, R, Forchhammer, K, Hartmann, M.D. | | Deposit date: | 2019-10-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Functional and structural characterization of PII-like protein CutA does not support involvement in heavy metal tolerance and hints at a small-molecule carrying/signaling role.
Febs J., 288, 2021
|
|
6T7E
 
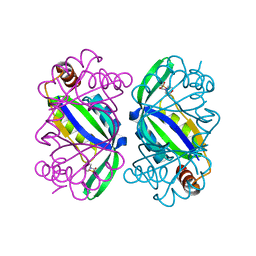 | | PII-like protein CutA from Nostoc sp. PCC7120 in complex with MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Periplasmic divalent cation tolerance protein | | Authors: | Selim, K.A, Albrecht, R, Forchhammer, K, Hartmann, M.D. | | Deposit date: | 2019-10-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Functional and structural characterization of PII-like protein CutA does not support involvement in heavy metal tolerance and hints at a small-molecule carrying/signaling role.
Febs J., 288, 2021
|
|
7E3U
 
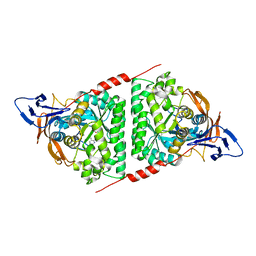 | | Crystal structure of the Pseudomonas aeruginosa dihydropyrimidinase complexed with 5-AU | | Descriptor: | 5-AMINO-1H-PYRIMIDINE-2,4-DIONE, D-hydantoinase/dihydropyrimidinase, ZINC ION | | Authors: | Yang, Y.C, Luo, R.H, Huang, Y.H, Huang, C.Y, Lin, E.S. | | Deposit date: | 2021-02-09 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.159 Å) | | Cite: | Molecular Insights into How the Dimetal Center in Dihydropyrimidinase Can Bind the Thymine Antagonist 5-Aminouracil: A Different Binding Mode from the Anticancer Drug 5-Fluorouracil.
Bioinorg Chem Appl, 2022, 2022
|
|
7E50
 
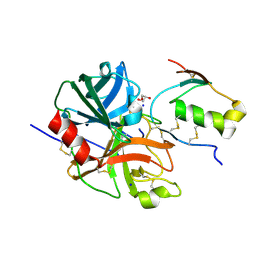 | | Crystal structure of human microplasmin in complex with kazal-type inhibitor AaTI | | Descriptor: | AAEL006007-PA, GLYCEROL, Plasminogen, ... | | Authors: | Varsha, A.W, Jobichen, C, Mok, Y.K. | | Deposit date: | 2021-02-16 | | Release date: | 2022-02-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of Aedes aegypti trypsin inhibitor in complex with mu-plasmin reveals role for scaffold stability in Kazal-type serine protease inhibitor.
Protein Sci., 31, 2022
|
|
6UE0
 
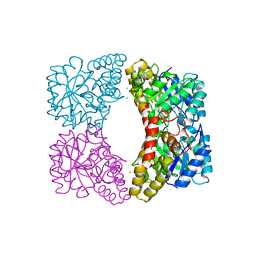 | | Crystal structure of dihydrodipicolinate synthase from Klebsiella pneumoniae bound to pyruvate | | Descriptor: | 4-hydroxy-tetrahydrodipicolinate synthase, CHLORIDE ION, SULFATE ION | | Authors: | Impey, R.E, Lee, M, Hawkins, D.A, Sutton, J.M, Panjikar, S, Perugini, M.A, Soares da Costa, T.P. | | Deposit date: | 2019-09-20 | | Release date: | 2020-02-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Mis-annotations of a promising antibiotic target in high-priority gram-negative pathogens.
Febs Lett., 594, 2020
|
|
6EX7
 
 | | Crystal structure of NDM-1 metallo-beta-lactamase in complex with Cd ions and a hydrolyzed beta-lactam ligand - new refinement | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, CADMIUM ION, ... | | Authors: | Kim, Y, Raczynska, J.E, Shabalin, I.G, Jaskolski, M, Minor, W, Wlodawer, A, Tesar, C, Jedrzejczak, R, Babnigg, J, Mire, J, Sacchettini, J, Joachimiak, A. | | Deposit date: | 2017-11-07 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
7F2N
 
 | |
5KK3
 
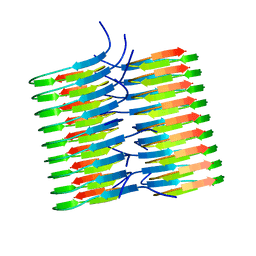 | | Atomic Resolution Structure of Monomorphic AB42 Amyloid Fibrils | | Descriptor: | Beta-amyloid protein 42 | | Authors: | Colvin, M.T, Silvers, R, Zhe Ni, Q, Can, T.V, Sergeyev, I, Rosay, M, Donovan, K.J, Michael, B, Wall, J, Linse, S, Griffin, R.G. | | Deposit date: | 2016-06-20 | | Release date: | 2016-07-13 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Atomic Resolution Structure of Monomorphic A beta 42 Amyloid Fibrils.
J.Am.Chem.Soc., 138, 2016
|
|
7F25
 
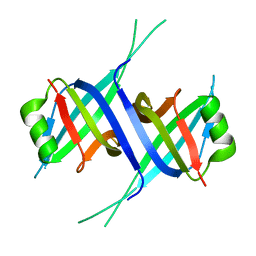 | |
5JO9
 
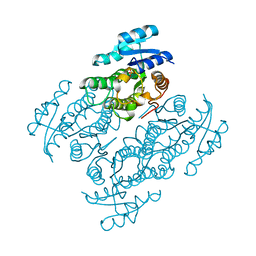 | | Structural characterization of the thermostable Bradyrhizobium japonicum d-sorbitol dehydrogenase | | Descriptor: | PHOSPHATE ION, Ribitol 2-dehydrogenase, sorbitol | | Authors: | Fredslund, F, Otten, H, Navarro Poulsen, J.-C, Lo Leggio, L. | | Deposit date: | 2016-05-02 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.894 Å) | | Cite: | Structural characterization of the thermostable Bradyrhizobium japonicumD-sorbitol dehydrogenase.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
6ISD
 
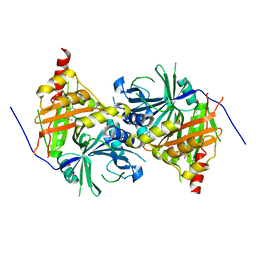 | | Crystal structure of Arabidopsis thaliana HPPD complexed with sulcotrione | | Descriptor: | 2-[2-chloro-4-(methylsulfonyl)benzoyl]cyclohexane-1,3-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Yang, W.C, Yang, G.F. | | Deposit date: | 2018-11-16 | | Release date: | 2018-11-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular insights into the mechanism of 4-hydroxyphenylpyruvate dioxygenase inhibition: enzyme kinetics, X-ray crystallography and computational simulations.
FEBS J., 286, 2019
|
|
6GKG
 
 | |
5J78
 
 | | Crystal structure of an Acetylating Aldehyde Dehydrogenase from Geobacillus thermoglucosidasius | | Descriptor: | ACETATE ION, Acetaldehyde dehydrogenase (Acetylating), GLYCEROL, ... | | Authors: | Crennell, S.J, Extance, J.P, Danson, M.J. | | Deposit date: | 2016-04-06 | | Release date: | 2016-09-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of an acetylating aldehyde dehydrogenase from the thermophilic ethanologen Geobacillus thermoglucosidasius.
Protein Sci., 25, 2016
|
|
7R0K
 
 | | Crystal structure of Polymerase I from phage G20c | | Descriptor: | DNA polymerase I | | Authors: | Welin, M, Svensson, A, Hakansson, M, Al-Karadaghi, S, Linares-Pasten, J.A, Jasilionis, A, Nordberg Karlsson, E, Ahlqvist, J. | | Deposit date: | 2022-02-02 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.972 Å) | | Cite: | Crystal structure of DNA polymerase I from Thermus phage G20c.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
6UX5
 
 | |
