8IE6
 
 | | Crystal structure of DAPK1 in complex with pinostilbene | | Descriptor: | 3-[(E)-2-(4-hydroxyphenyl)ethenyl]-5-methoxy-phenol, Death-associated protein kinase 1, SULFATE ION | | Authors: | Yokoyama, T. | | Deposit date: | 2023-02-15 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Characterization of the molecular interactions between resveratrol derivatives and death-associated protein kinase 1.
Febs J., 290, 2023
|
|
8IE7
 
 | | Crystal structure of DAPK1 in complex with pterostilbene | | Descriptor: | Death-associated protein kinase 1, Pterostilbene, SULFATE ION | | Authors: | Yokoyama, T. | | Deposit date: | 2023-02-15 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Characterization of the molecular interactions between resveratrol derivatives and death-associated protein kinase 1.
Febs J., 290, 2023
|
|
8IE5
 
 | | Crystal structure of DAPK1 in complex with oxyresveratrol | | Descriptor: | Death-associated protein kinase 1, SULFATE ION, trans-oxyresveratrol | | Authors: | Yokoyama, T. | | Deposit date: | 2023-02-15 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Characterization of the molecular interactions between resveratrol derivatives and death-associated protein kinase 1.
Febs J., 290, 2023
|
|
8SP5
 
 | |
6OFD
 
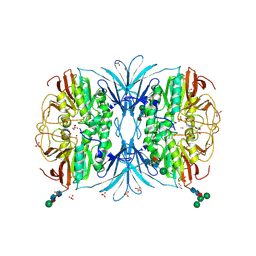 | | The crystal structure of octadecyloxy(naphthalen-1-yl)methylphosphonic acid in complex with red kidney bean purple acid phosphatase | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Feder, D, Schenk, G, Guddat, L.W, Hussein, W.M, McGeary, R.P, Kan, M.W. | | Deposit date: | 2019-03-29 | | Release date: | 2019-09-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Synthesis, evaluation and structural investigations of potent purple acid phosphatase inhibitors as drug leads for osteoporosis.
Eur.J.Med.Chem., 182, 2019
|
|
7N5O
 
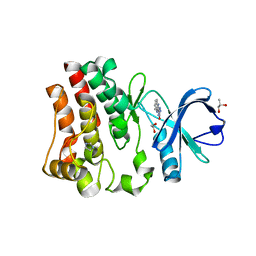 | | Fragment-Based Discovery of a Novel Bruton's Tyrosine Kinase Inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 5-(1H-benzimidazol-2-yl)-2,4-dihydro-3H-1,2,4-triazol-3-one, IMIDAZOLE, ... | | Authors: | Dougan, D.R, Lawson, J.D. | | Deposit date: | 2021-06-06 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Discovery of the Bruton's Tyrosine Kinase Inhibitor Clinical Candidate TAK-020 ( S )-5-(1-((1-Acryloylpyrrolidin-3-yl)oxy)isoquinolin-3-yl)-2,4-dihydro-3 H -1,2,4-triazol-3-one, by Fragment-Based Drug Design.
J.Med.Chem., 64, 2021
|
|
8B47
 
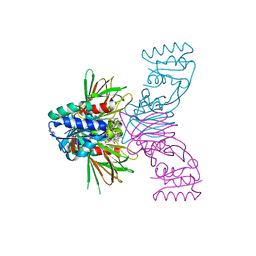 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a cyclic di-adenosine derivative | | Descriptor: | (1~{R},23~{R},24~{S},25~{R})-14-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl]-7-azanyl-24,25-bis(oxidanyl)-20,20-bis(oxidanylidene)-26-oxa-20$l^{6}-thia-2,4,6,9,14,17,21-heptazatetracyclo[21.2.1.0^{2,10}.0^{3,8}]hexacosa-3,5,7,9-tetraen-11-yn-16-one, NAD kinase 1, PENTAETHYLENE GLYCOL, ... | | Authors: | Gelin, M, Labesse, G. | | Deposit date: | 2022-09-20 | | Release date: | 2023-01-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Synthesis and structure-activity relationship studies of original cyclic diadenosine derivatives as nanomolar inhibitors of NAD kinase from pathogenic bacteria.
Eur.J.Med.Chem., 246, 2023
|
|
9EY4
 
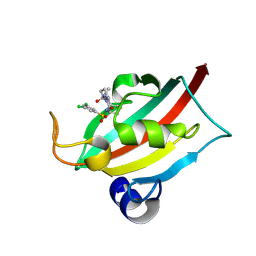 | | The FK1 domain of FKBP51 in complex with (3S,11S)-12-((3,5-dichlorophenyl)sulfonyl)-5-oxo-11-vinyldecahydro-1H-6,10-epiminopyrrolo[1,2-a]azonine-3-carboxamide | | Descriptor: | (1~{S},4~{S},7~{S},8~{S},9~{R})-13-[3,5-bis(chloranyl)phenyl]sulfonyl-8-ethenyl-2-oxidanylidene-3,13-diazatricyclo[7.3.1.0^{3,7}]tridecane-4-carboxamide, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Krajczy, P, Hausch, F. | | Deposit date: | 2024-04-09 | | Release date: | 2024-06-12 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Structure-Based Design of Ultrapotent Tricyclic Ligands for FK506-Binding Proteins.
Chemistry, 30, 2024
|
|
9EY3
 
 | | The FK1 domain of FKBP51 in complex with (3S,11S,11aS)-12-((3,5-dichlorophenyl)sulfonyl)-5-oxo-11-vinyldecahydro-1H-6,10-epiminopyrrolo[1,2-a]azonine-3-carboxylic acid | | Descriptor: | (1~{S},4~{S},7~{S},8~{S},9~{R})-13-[3,5-bis(chloranyl)phenyl]sulfonyl-8-ethenyl-2-oxidanylidene-3,13-diazatricyclo[7.3.1.0^{3,7}]tridecane-4-carboxylic acid, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Krajczy, P, Hausch, F. | | Deposit date: | 2024-04-09 | | Release date: | 2024-06-12 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Structure-Based Design of Ultrapotent Tricyclic Ligands for FK506-Binding Proteins.
Chemistry, 30, 2024
|
|
7MWT
 
 | |
6OF5
 
 | | The crystal structure of dodecyloxy(naphthalen-1-yl)methylphosphonic acid in complex with red kidney bean purple acid phosphatase | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, FE (III) ION, ... | | Authors: | Feder, D, Schenk, G, Guddat, L.W, Hussein, W.M, McGeary, R.P. | | Deposit date: | 2019-03-28 | | Release date: | 2019-09-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Synthesis, evaluation and structural investigations of potent purple acid phosphatase inhibitors as drug leads for osteoporosis.
Eur.J.Med.Chem., 182, 2019
|
|
7R0P
 
 | | CRYSTAL STRUCTURE OF E.coli ALCOHOL DEHYDROGENASE - FucO MUTANT F254I COMPLEXED WITH FE, NAD+, AND ETHYLENE GLYCOL | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, Lactaldehyde reductase, ... | | Authors: | Shruthi, S, Tiila, R.K, Rikkert, W, Mikael, W. | | Deposit date: | 2022-02-02 | | Release date: | 2022-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of lactaldehyde reductase, FucO, link enzyme activity to hydrogen bond networks and conformational dynamics.
Febs J., 290, 2023
|
|
7R5T
 
 | | CRYSTAL STRUCTURE OF E.coli ALCOHOL DEHYDROGENASE - FucO MUTANT F254I COMPLEXED WITH FE, NADH, AND GLYCEROL | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ADENOSINE-5-DIPHOSPHORIBOSE, FE (III) ION, ... | | Authors: | Sridhar, S, Kiema, T.R, Wierenga, R, Widersten, M. | | Deposit date: | 2022-02-11 | | Release date: | 2022-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures of lactaldehyde reductase, FucO, link enzyme activity to hydrogen bond networks and conformational dynamics.
Febs J., 290, 2023
|
|
9H1M
 
 | | Recombinant ferric horseradish peroxidase C1A | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DIHYDROGENPHOSPHATE ION, ... | | Authors: | Nesa, M.L, Mandal, S.K, Toelzer, C, Humer, D, Moody, P.C.E, Berger, I, Spadiut, O, Raven, E.L. | | Deposit date: | 2024-10-09 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal structure of ferric recombinant horseradish peroxidase.
J.Biol.Inorg.Chem., 30, 2025
|
|
7QBQ
 
 | | Human butyrylcholinesterase in complex with (Z)-N-benzyl-1-(8-hydroxyquinolin-2-yl)methanimine oxide | | Descriptor: | 1-(8-oxidanylquinolin-2-yl)-N-(phenylmethyl)methanimine oxide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Denic, M, Chioua, M, Knez, D, Gobec, S, Nachon, F, Marco-Contelles, J.L, Brazzolotto, X. | | Deposit date: | 2021-11-19 | | Release date: | 2022-11-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | 8-Hydroxyquinolylnitrones as multifunctional ligands for the therapy of neurodegenerative diseases.
Acta Pharm Sin B, 13, 2023
|
|
9INV
 
 | | Crystal structure of DAPK1 in complex with isoliquiritigenin | | Descriptor: | 2',4,4'-TRIHYDROXYCHALCONE, Death-associated protein kinase 1, SULFATE ION | | Authors: | Yokoyama, T. | | Deposit date: | 2024-07-08 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Discovery and optimization of isoliquiritigenin as a death-associated protein kinase 1 inhibitor.
Eur.J.Med.Chem., 279, 2024
|
|
9FZ6
 
 | | A 2.58A crystal structure of S. aureus DNA gyrase and DNA with metals identified through anomalous scattering | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*G)-3'), DNA (5'-D(*AP*GP*TP*AP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), ... | | Authors: | Morgan, H, Duman, R, Bax, B.D, Warren, A.J. | | Deposit date: | 2024-07-04 | | Release date: | 2024-10-23 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | How Do Gepotidacin and Zoliflodacin Stabilize DNA Cleavage Complexes with Bacterial Type IIA Topoisomerases? 1. Experimental Definition of Metal Binding Sites.
Int J Mol Sci, 25, 2024
|
|
8CRF
 
 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 5E11 refined against anomalous diffraction data | | Descriptor: | Host translation inhibitor nsp1, ~{N}-methyl-1-(4-thiophen-2-ylphenyl)methanamine | | Authors: | Ma, S, Mykhaylyk, V, Pinotsis, N, Bowler, M.W, Kozielski, F. | | Deposit date: | 2023-03-08 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | High-Confidence Placement of Fragments into Electron Density Using Anomalous Diffraction-A Case Study Using Hits Targeting SARS-CoV-2 Non-Structural Protein 1.
Int J Mol Sci, 24, 2023
|
|
7XZQ
 
 | | Crystal structure of TNIK-thiopeptide TP1 complex | | Descriptor: | 1,4-BUTANEDIOL, TRAF2 and NCK-interacting protein kinase, thiopeptide TP1 | | Authors: | Hamada, K, Vinogradov, A.A, Zhang, Y, Chang, J.S, Nishimura, H, Goto, Y, Onaka, H, Suga, H, Ogata, K, Sengoku, T. | | Deposit date: | 2022-06-03 | | Release date: | 2022-10-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | De Novo Discovery of Thiopeptide Pseudo-natural Products Acting as Potent and Selective TNIK Kinase Inhibitors.
J.Am.Chem.Soc., 144, 2022
|
|
6ZK0
 
 | | 1.47A human IMPase with ebselen | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Inositol monophosphatase 1, ... | | Authors: | Bax, B.D, Fenn, G.D. | | Deposit date: | 2020-06-29 | | Release date: | 2020-09-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystallization and structure of ebselen bound to Cys141 of human inositol monophosphatase.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
1FEH
 
 | | FE-ONLY HYDROGENASE FROM CLOSTRIDIUM PASTEURIANUM | | Descriptor: | 2 IRON/2 SULFUR/5 CARBONYL/2 WATER INORGANIC CLUSTER, FE2/S2 (INORGANIC) CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Peters, J.W, Lanzilotta, W.N, Lemon, B.J, Seefeldt, L.C. | | Deposit date: | 1998-10-28 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of the Fe-only hydrogenase (CpI) from Clostridium pasteurianum to 1.8 angstrom resolution.
Science, 282, 1998
|
|
8T5N
 
 | |
7MYC
 
 | |
8BR4
 
 | | Structure of GAPDH from Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Kumar, A, Karthikeyan, S. | | Deposit date: | 2022-11-22 | | Release date: | 2023-11-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Stoichiometry of ligand binding and role of C-terminal lysines in Mycobacterium tuberculosis and human GAPDH multifunctionality.
Febs J., 2024
|
|
8B2C
 
 | | Crystal structure of type I dehydroquinase from Salmonella typhi inhibited by an epoxide derivative | | Descriptor: | (1~{S},2~{R},4~{R},5~{S},6~{S})-2,4,5-trihydroxy-7-oxabicyclo[4.1.0]heptane-2-carboxylic acid, 3-dehydroquinate dehydratase, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Otero, J.M, Rodriguez, A, Maneiro, M, Lence, E, Thompson, P, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2022-09-13 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Quinate-based ligands for irreversible inactivation of the bacterial virulence factor DHQ1 enzyme-A molecular insight.
Front Mol Biosci, 10, 2023
|
|
