8JXW
 
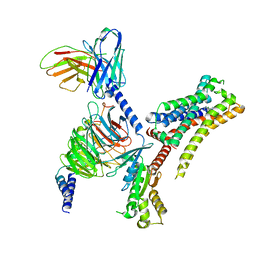 | | VUF6884-bound H4R/Gi complex | | Descriptor: | 2-chloranyl-6-(4-methylpiperazin-1-yl)benzo[b][1,4]benzoxazepine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | He, Y, Xia, R. | | Deposit date: | 2023-07-01 | | Release date: | 2024-03-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural basis of ligand recognition and design of antihistamines targeting histamine H 4 receptor.
Nat Commun, 15, 2024
|
|
8JXV
 
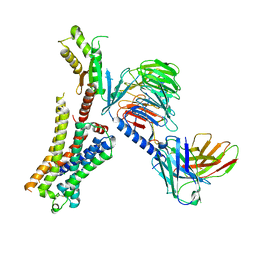 | | Clozapine-bound H4R/Gi complex | | Descriptor: | Clozapine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | He, Y, Xia, R. | | Deposit date: | 2023-07-01 | | Release date: | 2024-03-20 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural basis of ligand recognition and design of antihistamines targeting histamine H 4 receptor.
Nat Commun, 15, 2024
|
|
8JRN
 
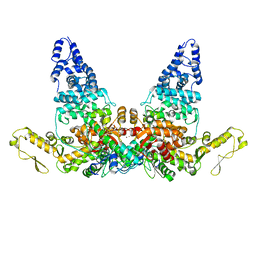 | | Structure of E6AP-E6 complex in Att1 state | | Descriptor: | Protein E6, Ubiquitin-protein ligase E3A, ZINC ION | | Authors: | Wang, Z, Yu, X. | | Deposit date: | 2023-06-17 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insights into the functional mechanism of the ubiquitin ligase E6AP.
Nat Commun, 15, 2024
|
|
8JRQ
 
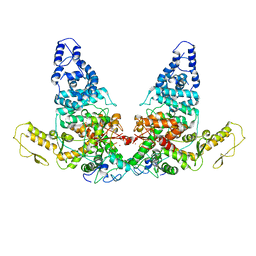 | | Structure of E6AP-E6 complex in Det1 state | | Descriptor: | Protein E6, Ubiquitin-protein ligase E3A, ZINC ION | | Authors: | Wang, Z, Yu, X. | | Deposit date: | 2023-06-17 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.15 Å) | | Cite: | Structural insights into the functional mechanism of the ubiquitin ligase E6AP.
Nat Commun, 15, 2024
|
|
8JRP
 
 | | Structure of E6AP-E6 complex in Att3 state | | Descriptor: | Protein E6, Ubiquitin-protein ligase E3A, ZINC ION | | Authors: | Wang, Z, Yu, X. | | Deposit date: | 2023-06-17 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structural insights into the functional mechanism of the ubiquitin ligase E6AP.
Nat Commun, 15, 2024
|
|
8JRR
 
 | | Structure of E6AP-E6 complex in Det2 state | | Descriptor: | Protein E6, Ubiquitin-protein ligase E3A, ZINC ION | | Authors: | Wang, Z, Yu, X. | | Deposit date: | 2023-06-17 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.35 Å) | | Cite: | Structural insights into the functional mechanism of the ubiquitin ligase E6AP.
Nat Commun, 15, 2024
|
|
8JRO
 
 | | Structure of E6AP-E6 complex in Att2 state | | Descriptor: | Protein E6, Ubiquitin-protein ligase E3A, ZINC ION | | Authors: | Wang, Z, Yu, X. | | Deposit date: | 2023-06-17 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural insights into the functional mechanism of the ubiquitin ligase E6AP.
Nat Commun, 15, 2024
|
|
9H9Y
 
 | | Crystal structure of metal-free LmrR_V15Bpy variant BVS in a closed state | | Descriptor: | Transcriptional regulator, PadR-like family | | Authors: | Thunnissen, A.M.W.H, Jiang, R, Casilli, F, Aalbers, F, Roelfes, G. | | Deposit date: | 2024-11-01 | | Release date: | 2025-03-05 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | An Artificial Copper-Michaelase Featuring a Genetically Encoded Bipyridine Ligand for Asymmetric Additions to Nitroalkenes.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9H9X
 
 | | Crystal structure of metal-free LmrR_V15Bpy in a closed state | | Descriptor: | Transcriptional regulator, PadR-like family | | Authors: | Thunnissen, A.M.W.H, Jiang, R, Casilli, F, Aalbers, F, Roelfes, G. | | Deposit date: | 2024-11-01 | | Release date: | 2025-03-05 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | An Artificial Copper-Michaelase Featuring a Genetically Encoded Bipyridine Ligand for Asymmetric Additions to Nitroalkenes.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9HA0
 
 | | Crystal structure of Cu(II)-bound LmrR_V15Bpy variant BVS | | Descriptor: | COPPER (II) ION, GLYCEROL, IMIDAZOLE, ... | | Authors: | Thunnissen, A.M.W.H, Jiang, R, Casilli, F, Aalbers, F, Roelfes, G. | | Deposit date: | 2024-11-01 | | Release date: | 2025-03-05 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | An Artificial Copper-Michaelase Featuring a Genetically Encoded Bipyridine Ligand for Asymmetric Additions to Nitroalkenes.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
1YT6
 
 | | NMR structure of peptide SD | | Descriptor: | peptide SD | | Authors: | Murata, T, Hemmi, H, Nakamura, S, Shimizu, K, Suzuki, Y, Yamaguchi, I. | | Deposit date: | 2005-02-10 | | Release date: | 2005-09-27 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Structure, epitope mapping, and docking simulation of a gibberellin mimic peptide as a peptidyl mimotope for a hydrophobic ligand.
Febs J., 272, 2005
|
|
7ONP
 
 | | Wild type carbonic anhydrase II with bound IrCp* complex to generate an artificial transfer hydrogenase (ATHase) | | Descriptor: | 4-[2-(9-chloranyl-2',3',4',5',6'-pentamethyl-4-nitro-7-oxidanylidene-spiro[1$l^{4},8-diaza-9$l^{8}-iridabicyclo[4.3.0]nona-1,3,5-triene-9,1'-1$l^{8}-iridapentacyclo[2.2.0.0^{1,3}.0^{1,5}.0^{2,6}]hexane]-8-yl)ethyl]benzenesulfonamide, ACETATE ION, Carbonic anhydrase 2, ... | | Authors: | Stein, A, Dongping, C, Cotelle, Y, Rebelein, J.G, Ward, T.R. | | Deposit date: | 2021-05-25 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.408 Å) | | Cite: | A Dual Anchoring Strategy for the Directed Evolution of Improved Artificial Transfer Hydrogenases Based on Carbonic Anhydrase.
Acs Cent.Sci., 7, 2021
|
|
8OQ6
 
 | | CryoEM structure of human rho1 GABAA receptor apo state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, DECANE, ... | | Authors: | Chen, F, Victor, T, John, C, Rebecca, J.H, Lindahl, E. | | Deposit date: | 2023-04-11 | | Release date: | 2023-08-30 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structure and dynamics of differential ligand binding in the human rho-type GABA A receptor.
Neuron, 111, 2023
|
|
3IE9
 
 | | Structure of oxidized M98L mutant of amicyanin | | Descriptor: | ACETATE ION, Amicyanin, CHLORIDE ION, ... | | Authors: | Sukumar, N, Davidson, V.L. | | Deposit date: | 2009-07-22 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Defining the role of the axial ligand of the type 1 copper site in amicyanin by replacement of methionine with leucine.
Biochemistry, 48, 2009
|
|
8OQA
 
 | | CryoEM structure of human rho1 GABAA receptor in complex with GABA and picrotoxin | | Descriptor: | (1aR,2aR,3S,6R,6aS,8aS,8bR,9R)-2a-hydroxy-8b-methyl-9-(prop-1-en-2-yl)hexahydro-3,6-methano-1,5,7-trioxacyclopenta[ij]c yclopropa[a]azulene-4,8(3H)-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Chen, F, Victor, T, John, C, Rebecca, J.H, Lindahl, E. | | Deposit date: | 2023-04-11 | | Release date: | 2023-08-30 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure and dynamics of differential ligand binding in the human rho-type GABA A receptor.
Neuron, 111, 2023
|
|
8OQ7
 
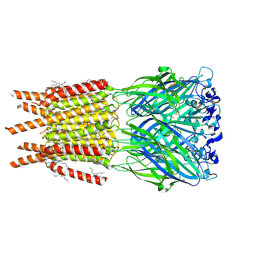 | | CryoEM structure of human rho1 GABAA receptor in complex with inhibitor TPMPA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, DECANE, ... | | Authors: | Chen, F, Victor, T, John, C, Rebecca, J.H, Lindahl, E. | | Deposit date: | 2023-04-11 | | Release date: | 2023-08-30 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Structure and dynamics of differential ligand binding in the human rho-type GABA A receptor.
Neuron, 111, 2023
|
|
8OQ8
 
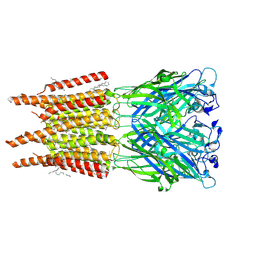 | | CryoEM structure of human rho1 GABAA receptor in complex with pore blocker picrotoxin | | Descriptor: | (1aR,2aR,3S,6R,6aS,8aS,8bR,9R)-2a-hydroxy-8b-methyl-9-(prop-1-en-2-yl)hexahydro-3,6-methano-1,5,7-trioxacyclopenta[ij]c yclopropa[a]azulene-4,8(3H)-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, DECANE, ... | | Authors: | Chen, F, Victor, T, John, C, Rebecca, J.H, Lindahl, E. | | Deposit date: | 2023-04-11 | | Release date: | 2023-08-30 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure and dynamics of differential ligand binding in the human rho-type GABA A receptor.
Neuron, 111, 2023
|
|
8OP9
 
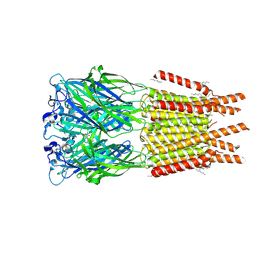 | | CryoEM structure of human rho1 GABAA receptor in complex with GABA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Chen, F, Victor, T, John, C, Rebecca, J.H, Lindahl, E. | | Deposit date: | 2023-04-06 | | Release date: | 2023-08-30 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structure and dynamics of differential ligand binding in the human rho-type GABA A receptor.
Neuron, 111, 2023
|
|
5ZWH
 
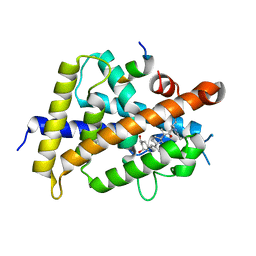 | | Covalent bond formation between histidine of Vitamin D receptor (VDR) and a full agonist having an ene-ynone group via conjugate addition reaction | | Descriptor: | (2S)-2-[(1R,3aS,4E,7aR)-7a-methyl-4-[2-[(3R,5R)-4-methylidene-3,5-bis(oxidanyl)cyclohexylidene]ethylidene]-2,3,3a,5,6,7-hexahydro-1H-inden-1-yl]oct-4,6-diene-3-one, (E,2S)-2-[(1R,3aS,4E,7aR)-7a-methyl-4-[2-[(3R,5R)-4-methylidene-3,5-bis(oxidanyl)cyclohexylidene]ethylidene]-2,3,3a,5,6,7-hexahydro-1H-inden-1-yl]oct-6-en-4-yn-3-one, 13-meric peptide from DRIP205 NR2 BOX peptide, ... | | Authors: | Yoshizawa, M, Itoh, T, Anami, Y, Kato, A, Yoshimoto, N, Yamamoto, K. | | Deposit date: | 2018-05-15 | | Release date: | 2018-07-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Identification of the Histidine Residue in Vitamin D Receptor That Covalently Binds to Electrophilic Ligands
J. Med. Chem., 61, 2018
|
|
7AH4
 
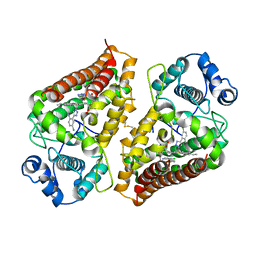 | | Crystal structure of indoleamine 2,3-dioxygenase 1 (IDO1) in complex with ferric heme and MMG-0363 | | Descriptor: | 4-chloranyl-2-(2~{H}-1,2,3-triazol-4-yl)aniline, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Roehrig, U.F, Reynaud, A, Pojer, F, Michielin, O, Zoete, V. | | Deposit date: | 2020-09-24 | | Release date: | 2021-02-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Azole-Based Indoleamine 2,3-Dioxygenase 1 (IDO1) Inhibitors.
J.Med.Chem., 64, 2021
|
|
7AH6
 
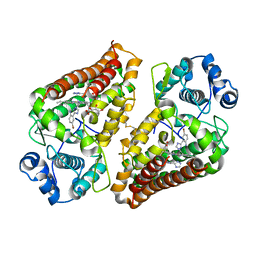 | | Crystal structure of indoleamine 2,3-dioxygenase 1 (IDO1) in complex with ferric heme and MMG-0752 | | Descriptor: | 4-bromanyl-2-(4~{H}-1,2,4-triazol-3-yl)aniline, GLYCEROL, Indoleamine 2,3-dioxygenase 1, ... | | Authors: | Roehrig, U.F, Reynaud, A, Pojer, F, Michielin, O, Zoete, V. | | Deposit date: | 2020-09-24 | | Release date: | 2021-02-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.998 Å) | | Cite: | Azole-Based Indoleamine 2,3-Dioxygenase 1 (IDO1) Inhibitors.
J.Med.Chem., 64, 2021
|
|
7AH5
 
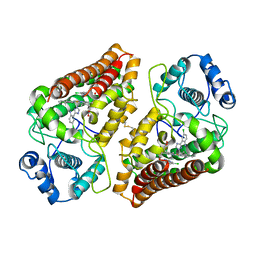 | | Crystal structure of indoleamine 2,3-dioxygenase 1 (IDO1) in complex with ferric heme and MMG-0706 | | Descriptor: | 4-chloranyl-2-(1~{H}-1,2,4-triazol-5-yl)aniline, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Roehrig, U.F, Reynaud, A, Pojer, F, Michielin, O, Zoete, V. | | Deposit date: | 2020-09-24 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Azole-Based Indoleamine 2,3-Dioxygenase 1 (IDO1) Inhibitors.
J.Med.Chem., 64, 2021
|
|
9EAJ
 
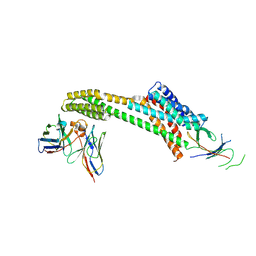 | |
4APU
 
 | | PR X-Ray structures in agonist conformations reveal two different mechanisms for partial agonism in 11beta-substituted steroids | | Descriptor: | (8S,11R,13S,14S,16S,17S)-17-cyclopropylcarbonyl-16-ethenyl-13-methyl-11-(4-pyridin-3-ylphenyl)-2,6,7,8,11,12,14,15,16,17-decahydro-1H-cyclopenta[a]phenanthren-3-one, 2-CHLORO-N-[[4-(3,5-DIMETHYLISOXAZOL-4-YL)PHENYL]METHYL]-1,4-DIMETHYL-1H-PYRAZOLE-4-SULFONAMIDE, PROGESTERONE RECEPTOR, ... | | Authors: | Lusher, S.J, Raaijmakers, H.C.A, Bosch, R, Vu-Pham, D, McGuire, R, Oubrie, A, de Vlieg, J. | | Deposit date: | 2012-04-06 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structures of progesterone receptor ligand binding domain in its agonist state reveal differing mechanisms for mixed profiles of 11 beta-substituted steroids.
J. Biol. Chem., 287, 2012
|
|
9EAI
 
 | | Structure of nanobody AT206 in complex with the losartan-bound angiotensin II type I receptor (AT1R) | | Descriptor: | BAG2 Anti-BRIL Fab Heavy Chain, BAG2 Anti-BRIL Fab Light Chain, CHOLESTEROL, ... | | Authors: | Skiba, M.A, Liu, J, Kruse, A.C. | | Deposit date: | 2024-11-11 | | Release date: | 2025-03-05 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Epitope-directed selection of GPCR nanobody ligands with evolvable function.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
