3TR2
 
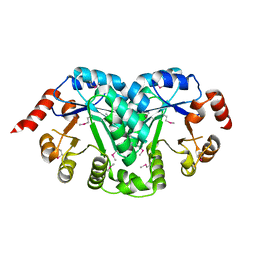 | | Structure of a orotidine 5'-phosphate decarboxylase (pyrF) from Coxiella burnetii | | Descriptor: | Orotidine 5'-phosphate decarboxylase | | Authors: | Cheung, J, Franklin, M, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
4GAY
 
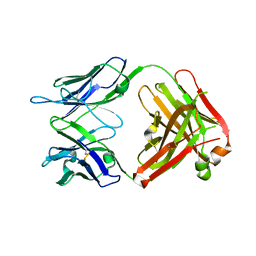 | | Structure of the broadly neutralizing antibody AP33 | | Descriptor: | NEUTRALIZING ANTIBODY AP33 HEAVY CHAIN, NEUTRALIZING ANTIBODY AP33 LIGHT CHAIN, TRIETHYLENE GLYCOL | | Authors: | Potter, J.A, Owsianka, A, Jeffery, N, Matthews, D, Keck, Z, Lau, P, Foung, S.K.H, Taylor, G.L, Patel, A.H. | | Deposit date: | 2012-07-26 | | Release date: | 2012-10-10 | | Last modified: | 2012-11-21 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Toward a Hepatitis C Virus Vaccine: the Structural Basis of Hepatitis C Virus Neutralization by AP33, a Broadly Neutralizing Antibody.
J.Virol., 86, 2012
|
|
6N2S
 
 | |
3QPX
 
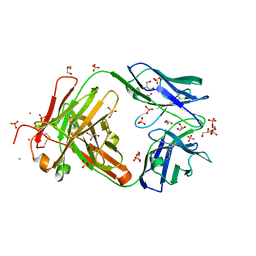 | | Crystal structure of Fab C2507 | | Descriptor: | CADMIUM ION, CHLORIDE ION, Fab C2507 heavy chain, ... | | Authors: | Luo, J, Teplyakov, A, Obmolova, O, Malia, T, Gilliland, G.L. | | Deposit date: | 2011-02-14 | | Release date: | 2011-08-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Fab C2507
To be Published
|
|
4G3P
 
 | |
4GHT
 
 | | Crystal structure of EV71 3C proteinase in complex with AG7088 | | Descriptor: | 3C proteinase, 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER | | Authors: | Chen, C, Wu, C, Cai, Q, Li, N, Peng, X, Cai, Y, Yin, K, Chen, X, Wang, X, Zhang, R, Liu, L, Chen, S, Li, J, Lin, T. | | Deposit date: | 2012-08-08 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structures of Enterovirus 71 3C proteinase (strain E2004104-TW-CDC) and its complex with rupintrivir
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4C1I
 
 | | Selective Inhibitors of PDE2, PDE9, and PDE10: Modulators of Activity of the Central Nervous System | | Descriptor: | (2S,3R)-3-(6-amino-9H-purin-9-yl)nonan-2-ol, CGMP-DEPENDENT 3', 5'-CYCLIC PHOSPHODIESTERASE, ... | | Authors: | Jorgensen, M, Kehler, J, Langgard, M, Svenstrup, N, Tagmose, L. | | Deposit date: | 2013-08-12 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Chapter 4: Selective Inhibitors of Pde2, Pde9, and Pde10: Modulators of Activity of the Central Nervous System
To be Published
|
|
4G3S
 
 | |
4G57
 
 | | Staphylococcal Nuclease double mutant I72L, I92L | | Descriptor: | Thermonuclease | | Authors: | Sakon, J, Stites, W.E, Gill, E, Latimer, E, Rosser, J. | | Deposit date: | 2012-07-17 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: |
|
|
4G61
 
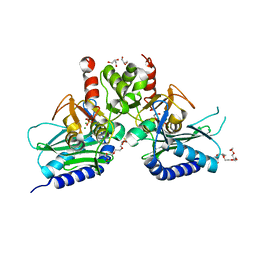 | | Crystal structure of IMPase/NADP phosphatase complexed with Mg2+ and phosphate | | Descriptor: | 1-HYDROXYSULFANYL-4-MERCAPTO-BUTANE-2,3-DIOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CHLORIDE ION, ... | | Authors: | Bhattacharyya, S, Dutta, D, Ghosh, A.K, Das, A.K. | | Deposit date: | 2012-07-18 | | Release date: | 2013-07-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural elucidation of the binding site and mode of inhibition of Li(+) and Mg(2+) in inositol monophosphatase.
Febs J., 281, 2014
|
|
2KOM
 
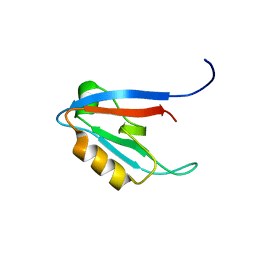 | | Solution structure of humar Par-3b PDZ2 (residues 451-549) | | Descriptor: | Partitioning defective 3 homolog | | Authors: | Volkman, B.F, Tyler, R.C, Peterson, F.C, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2009-09-24 | | Release date: | 2009-11-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Rapid, robotic, small-scale protein production for NMR screening and structure determination.
Protein Sci., 19, 2010
|
|
4G87
 
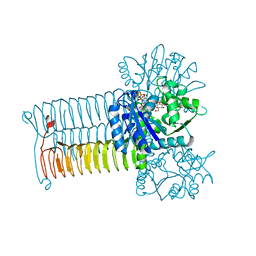 | | Crystal structure of GLMU from Mycobacterium tuberculosis snapshot 1 | | Descriptor: | Bifunctional protein GlmU, COBALT (II) ION, MAGNESIUM ION, ... | | Authors: | Jagtap, P.A, Verma, S.K, Prakash, B. | | Deposit date: | 2012-07-22 | | Release date: | 2013-03-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structures identify an atypical two-metal-ion mechanism for uridyltransfer in GlmU: its significance to sugar nucleotidyl transferases
J.Mol.Biol., 425, 2013
|
|
4C82
 
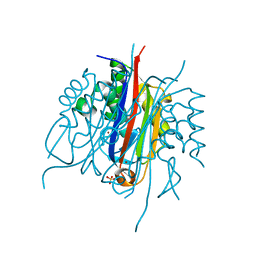 | | IspF (Plasmodium falciparum) unliganded structure | | Descriptor: | 2-C-METHYL-D-ERYTHRITOL 2,4-CYCLODIPHOSPHATE SYNTHASE, SULFATE ION, ZINC ION | | Authors: | O'Rourke, P.E.F, Kalinowska-Tluscik, J, Fyfe, P.K, Dawson, A, Hunter, W.N. | | Deposit date: | 2013-09-27 | | Release date: | 2014-01-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Ispf from Plasmodium Falciparum and Burkholderia Cenocepacia: Comparisons Inform Antimicrobial Drug Target Assessment.
Bmc Struct.Biol., 14, 2014
|
|
4GE0
 
 | | Schizosaccharomyces pombe DJ-1 T114P mutant | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Uncharacterized protein C22E12.03c | | Authors: | Madzelan, P, Labunska, T, Wilson, M.A. | | Deposit date: | 2012-08-01 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Influence of peptide dipoles and hydrogen bonds on reactive cysteine pK(a) values in fission yeast DJ-1.
Febs J., 279, 2012
|
|
4C8E
 
 | | IspF (Burkholderia cenocepacia) 2CMP complex | | Descriptor: | 2-C-METHYL-D-ERYTHRITOL 2,4-CYCLODIPHOSPHATE SYNTHASE, CYTIDINE-5'-MONOPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | O'Rourke, P.E.F, Kalinowska-Tluscik, J, Fyfe, P.K, Dawson, A, Hunter, W.N. | | Deposit date: | 2013-09-30 | | Release date: | 2014-01-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of Ispf from Plasmodium Falciparum and Burkholderia Cenocepacia: Comparisons Inform Antimicrobial Drug Target Assessment.
Bmc Struct.Biol., 14, 2014
|
|
4EWA
 
 | | Study on structure and function relationships in human Pirin with Fe ion | | Descriptor: | FE (III) ION, Pirin | | Authors: | Liu, F, Rehmani, I, Chen, L, Fu, R, Serrano, V, Wilson, D.W, Liu, A. | | Deposit date: | 2012-04-26 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Pirin is an iron-dependent redox regulator of NF-kappa B.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6NXZ
 
 | |
3RH5
 
 | | DNA Polymerase Beta Mutant (Y271) with a dideoxy-terminated primer with an incoming deoxynucleotide (dCTP) | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*CP*CP*GP*AP*CP*GP*CP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*(DDG))-3', ... | | Authors: | Cavanaugh, N.A, Beard, W.A, Batra, V.K, Perera, L, Pedersen, L.G, Wilson, S.H. | | Deposit date: | 2011-04-11 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Molecular insights into DNA polymerase deterrents for ribonucleotide insertion.
J.Biol.Chem., 286, 2011
|
|
4IK5
 
 | | High resolution structure of Delta-REST-GCaMP3 | | Descriptor: | CALCIUM ION, RCaMP, Green fluorescent protein | | Authors: | Chen, Y, Song, X, Miao, L, Zhu, Y, Ji, G. | | Deposit date: | 2012-12-25 | | Release date: | 2014-01-29 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insight into enhanced calcium indicator GCaMP3 and GCaMPJ to promote further improvement.
Protein Cell, 4, 2013
|
|
4ER8
 
 | | Structure of the REP associates tyrosine transposase bound to a REP hairpin | | Descriptor: | DNA (32-MER), NICKEL (II) ION, TnpArep for protein | | Authors: | Messing, S.A.J, Ton-Hoang, B, Hickman, A.B, Ghirlando, R, Chandler, M, Dyda, F. | | Deposit date: | 2012-04-19 | | Release date: | 2012-08-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The processing of repetitive extragenic palindromes: the structure of a repetitive extragenic palindrome bound to its associated nuclease.
Nucleic Acids Res., 40, 2012
|
|
3TRB
 
 | | Structure of an addiction module antidote protein of a HigA (higA) family from Coxiella burnetii | | Descriptor: | Virulence-associated protein I | | Authors: | Cheung, J, Franklin, M.C, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
4ID8
 
 | | The crystal structure of a [3Fe-4S] ferredoxin associated with CYP194A4 from R. palustris HaA2 | | Descriptor: | FE3-S4 CLUSTER, Putative ferredoxin | | Authors: | Zhou, W.H, Zhang, T, Zhang, A.L, Bell, S.G, Wong, L.-L. | | Deposit date: | 2012-12-11 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structure of a novel electron-transfer ferredoxin from Rhodopseudomonas palustris HaA2 which contains a histidine residue in its iron-sulfur cluster-binding motif.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3TQZ
 
 | | Structure of a deoxyuridine 5'-triphosphate nucleotidohydrolase (dut) from Coxiella burnetii | | Descriptor: | Deoxyuridine 5'-triphosphate nucleotidohydrolase, SULFATE ION | | Authors: | Cheung, J, Franklin, M, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
3TR6
 
 | | Structure of a O-methyltransferase from Coxiella burnetii | | Descriptor: | NICKEL (II) ION, O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Cheung, J, Franklin, M.C, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-28 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
4F0H
 
 | | UNACTIVATED RUBISCO with OXYGEN BOUND | | Descriptor: | OXYGEN MOLECULE, PHOSPHATE ION, Ribulose bisphosphate carboxylase large chain, ... | | Authors: | Stec, B. | | Deposit date: | 2012-05-04 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural mechanism of RuBisCO activation by carbamylation of the active site lysine.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
