1FJN
 
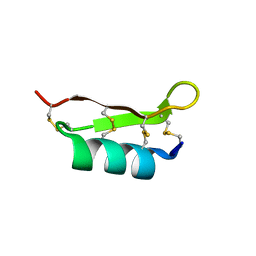 | |
1UNC
 
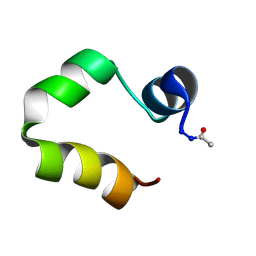 | | Solution structure of the human villin C-terminal headpiece subdomain | | Descriptor: | VILLIN 1 | | Authors: | Vermeulen, W, Van Troys, M, Vanhaesebrouck, P, Verschueren, M, Fant, F, Ampe, C, Martins, J, Borremans, F. | | Deposit date: | 2003-09-09 | | Release date: | 2004-07-15 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of the C-Terminal Headpiece Subdomains of Human Villin and Advillin, Evaluation of Headpiece F-Actin-Binding Requirements
Protein Sci., 13, 2004
|
|
1RW2
 
 | | Three-dimensional structure of Ku80 CTD | | Descriptor: | ATP-dependent DNA helicase II, 80 kDa subunit | | Authors: | Zhang, Z, Hu, W, Cano, L, Lee, T.D, Chen, D.J, Chen, Y. | | Deposit date: | 2003-12-15 | | Release date: | 2003-12-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal domain of Ku80 suggests important sites for protein-protein interactions.
STRUCTURE, 12, 2004
|
|
1FV7
 
 | | A TWO B-Z JUNCTION CONTAINING DNA RESOLVES INTO AN ALL RIGHT HANDED DOUBLE HELIX | | Descriptor: | 5'-D(*(5CM)P*GP*(5CM)P*GP*(0DC)P*(0DG)P*(5CM)P*GP*(5CM)P*G)-3' | | Authors: | Mauffret, O, El Amri, C, Santamaria, F, Tevanian, G, Rayner, B, Fermandjian, S. | | Deposit date: | 2000-09-19 | | Release date: | 2000-10-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A two B-Z junction containing DNA resolves into an all right-handed double-helix.
Nucleic Acids Res., 28, 2000
|
|
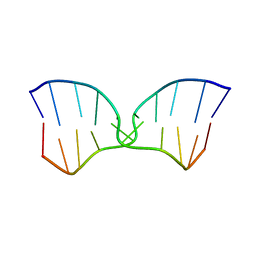
1FW5
 
 | | SOLUTION STRUCTURE OF MEMBRANE BINDING PEPTIDE OF SEMLIKI FOREST VIRUS MRNA CAPPING ENZYME NSP1 | | Descriptor: | NONSTRUCTURAL PROTEIN NSP1 | | Authors: | Lampio, A, Kilpelinen, I, Pesonen, S, Karhi, K, Auvinen, P, Somerharju, P, Kriinen, L. | | Deposit date: | 2000-09-21 | | Release date: | 2001-09-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Membrane binding mechanism of an RNA virus-capping enzyme.
J.Biol.Chem., 275, 2001
|
|
1FWQ
 
 | |
1H8C
 
 | | UBX domain from human faf1 | | Descriptor: | FAS-ASSOCIATED FACTOR 1 | | Authors: | Bycroft, M.M. | | Deposit date: | 2001-02-01 | | Release date: | 2001-02-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Ubx Domain: A Widespread Ubiquitin-Like Module
J.Mol.Biol., 307, 2001
|
|
1TRF
 
 | |
1DZ7
 
 | | Solution structure of the a-subunit of human chorionic gonadotropin [modeled without carbohydrate residues] | | Descriptor: | CHORIONIC GONADOTROPIN | | Authors: | Erbel, P.J.A, Karimi-Nejad, Y, De Beer, T, Boelens, R, Kamerling, J.P, Vliegenthart, J.F.G. | | Deposit date: | 2000-02-18 | | Release date: | 2000-02-29 | | Last modified: | 2019-10-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the alpha-subunit of human chorionic gonadotropin.
Eur.J.Biochem., 260, 1999
|
|
1UND
 
 | | Solution structure of the human advillin C-terminal headpiece subdomain | | Descriptor: | ADVILLIN | | Authors: | Vermeulen, W, Van Troys, M, Vanhaesebrouck, P, Verschueren, M, Fant, F, Ampe, C, Martins, J, Borremans, F. | | Deposit date: | 2003-09-09 | | Release date: | 2004-07-15 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of the C-Terminal Headpiece Subdomains of Human Villin and Advillin, Evaluation of Headpiece F-Actin-Binding Requirements
Protein Sci., 13, 2004
|
|
1DO9
 
 | | SOLUTION STRUCTURE OF OXIDIZED MICROSOMAL RABBIT CYTOCHROME B5. FACTORS DETERMINING THE HETEROGENEOUS BINDING OF THE HEME. | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Banci, L, Bertini, I, Rosato, A, Scacchieri, S. | | Deposit date: | 1999-12-20 | | Release date: | 2000-01-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of oxidized microsomal rabbit cytochrome b5. Factors determining the heterogeneous binding of the heme.
Eur.J.Biochem., 267, 2000
|
|
1SLM
 
 | |
1SLN
 
 | |
1GE9
 
 | | SOLUTION STRUCTURE OF THE RIBOSOME RECYCLING FACTOR | | Descriptor: | RIBOSOME RECYCLING FACTOR | | Authors: | Yoshida, T, Uchiyama, S, Nakano, H, Kashimori, H, Kijima, H, Ohshima, T, Saihara, Y, Ishino, T, Shimahara, T, Yoshida, T, Yokose, K, Ohkubo, T, Kaji, A, Kobayashi, Y. | | Deposit date: | 2000-10-19 | | Release date: | 2001-05-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the ribosome recycling factor from Aquifex aeolicus.
Biochemistry, 40, 2001
|
|
1GJE
 
 | | Peptide Antagonist of IGFBP-1, Minimized Average Structure | | Descriptor: | IGFBP-1 antagonist | | Authors: | Lowman, H.B, Chen, Y.M, Skelton, N.J, Mortensen, D.L, Tomlinson, E.E, Sadick, M.D, Robinson, I.C, Clark, R.G. | | Deposit date: | 2001-05-11 | | Release date: | 2001-05-30 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of a phage display-derived peptide that binds to insulin-like growth factor binding protein 1.
Biochemistry, 40, 2001
|
|
1GK5
 
 | | Solution Structure the mEGF/TGFalpha44-50 chimeric growth factor | | Descriptor: | Pro-epidermal growth factor,Protransforming growth factor alpha | | Authors: | Chamberlin, S.G, Brennan, L, Puddicombe, S.M, Davies, D.E, Turner, D.L. | | Deposit date: | 2001-08-08 | | Release date: | 2002-08-08 | | Last modified: | 2018-03-28 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Megf/Tgfalpha44-50 Chimeric Growth Factor.
Eur.J.Biochem., 268, 2001
|
|
1GEA
 
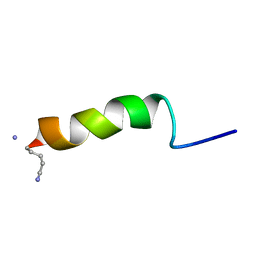 | | RECEPTOR-BOUND CONFORMATION OF PACAP21 | | Descriptor: | PITUITARY ADENYLATE CYCLASE ACTIVATING POLYPEPTIDE | | Authors: | Inooka, H, Ohtaki, T, Kitahara, O, Ikegami, T, Endo, S, Kitada, C, Ogi, K, Onda, H, Fujino, M, Shirakawa, M. | | Deposit date: | 2000-10-20 | | Release date: | 2001-04-20 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Conformation of a peptide ligand bound to its G-protein coupled receptor.
Nat.Struct.Biol., 8, 2001
|
|
1GJF
 
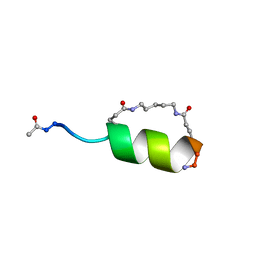 | | Peptide Antagonist of IGFBP1, (i,i+7) Covalently Restrained Analog, Minimized Average Structure | | Descriptor: | IGFBP-1 antagonist, PENTANE | | Authors: | Skelton, N.J, Chen, Y.M, Dubree, N, Quan, C, Jackson, D.Y, Cochran, A.G, Zobel, K, Deshayes, K, Baca, M, Pisabarro, M.T, Lowman, H.B. | | Deposit date: | 2001-05-11 | | Release date: | 2001-05-30 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of a phage display-derived peptide that binds to insulin-like growth factor binding protein 1.
Biochemistry, 40, 2001
|
|
1G26
 
 | |
1G91
 
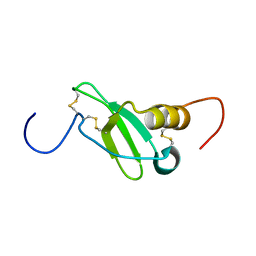 | | SOLUTION STRUCTURE OF MYELOID PROGENITOR INHIBITORY FACTOR-1 (MPIF-1) | | Descriptor: | MYELOID PROGENITOR INHIBITORY FACTOR-1 | | Authors: | Rajarathnam, K, Li, Y, Rohrer, T, Gentz, R. | | Deposit date: | 2000-11-21 | | Release date: | 2001-03-07 | | Last modified: | 2022-12-21 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of myeloid progenitor inhibitory factor-1 (MPIF-1), a novel monomeric CC chemokine.
J.Biol.Chem., 276, 2001
|
|
1EXG
 
 | | SOLUTION STRUCTURE OF A CELLULOSE BINDING DOMAIN FROM CELLULOMONAS FIMI BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | Descriptor: | EXO-1,4-BETA-D-GLYCANASE | | Authors: | Xu, G.-Y, Ong, E, Gilkes, N.R, Kilburn, D.G, Muhandiram, D.R, Harris-Brandts, M, Carver, J.P, Kay, L.E, Harvey, T.S. | | Deposit date: | 1995-03-14 | | Release date: | 1995-06-03 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a cellulose-binding domain from Cellulomonas fimi by nuclear magnetic resonance spectroscopy.
Biochemistry, 34, 1995
|
|
1VKR
 
 | |
1EMX
 
 | | SOLUTION STRUCTURE OF HPTX2, A TOXIN FROM HETEROPODA VENATORIA SPIDER VENOM THAT BLOCKS KV4.2 POTASSIUM CHANNEL | | Descriptor: | HETEROPODATOXIN 2 | | Authors: | Bernard, C, Legros, C, Ferrat, G, Bishoff, U, Marquardt, A, Pongs, O, Darbon, H. | | Deposit date: | 2000-03-20 | | Release date: | 2001-01-24 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of hpTX2, a toxin from Heteropoda venatoria spider that blocks Kv4.2 potassium channel.
Protein Sci., 9, 2000
|
|
1XHP
 
 | |
2N9L
 
 | | 1H, 13C, and 15N Chemical Shift Assignments for in-cell GB1 | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Ikeya, T, Hanashima, T, Hosoya, S, Shimazaki, M, Ikeda, S, Mishima, M, Guentert, P, Ito, Y. | | Deposit date: | 2015-11-30 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Improved in-cell structure determination of proteins at near-physiological concentration
Sci Rep, 6, 2016
|
|
