8BVW
 
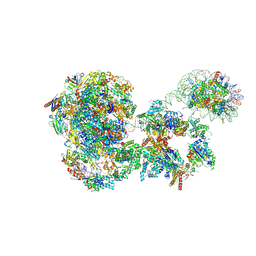 | | RNA polymerase II pre-initiation complex with the distal +1 nucleosome (PIC-Nuc18W) | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Abril-Garrido, J, Dienemann, C, Grabbe, F, Velychko, T, Lidschreiber, M, Wang, H, Cramer, P. | | Deposit date: | 2022-12-20 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of transcription reduction by a promoter-proximal +1 nucleosome.
Mol.Cell, 83, 2023
|
|
8C79
 
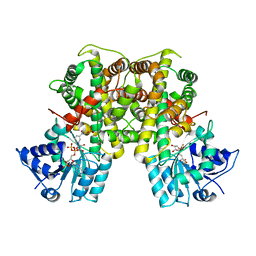 | | Crystal structure of Leishmania donovani 6-Phosphogluconate Dehydrogenase complexed with NADPH | | Descriptor: | 6-phosphogluconate dehydrogenase, decarboxylating, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Fritz-Wolf, K, Berneburg, I, Rahlfs, S, Becker, K. | | Deposit date: | 2023-01-13 | | Release date: | 2023-05-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of Leishmania donovani 6-Phosphogluconate Dehydrogenase and Inhibition by Phosphine Gold(I) Complexes: A Potential Approach to Leishmaniasis Treatment.
Int J Mol Sci, 24, 2023
|
|
4XZC
 
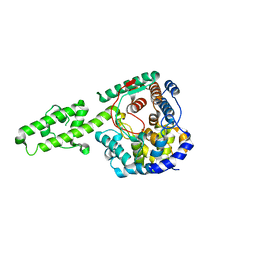 | | The crystal structure of Kupe virus nucleoprotein | | Descriptor: | Nucleoprotein | | Authors: | Guo, Y, Wang, W, Liu, X, Wang, X, Wang, J, Huo, T, Liu, B. | | Deposit date: | 2015-02-04 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structural and Functional Diversity of Nairovirus-Encoded Nucleoproteins.
J.Virol., 89, 2015
|
|
5L6S
 
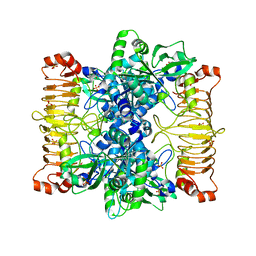 | | Crystal structure of E. coli ADP-glucose pyrophosphorylase (AGPase) in complex with a positive allosteric regulator beta-fructose-1,6-diphosphate (FBP) - AGPase*FBP | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, Glucose-1-phosphate adenylyltransferase, SULFATE ION | | Authors: | Cifuente, J.O, Albesa-Jove, D, Comino, N, Madariaga-Marcos, J, Agirre, J, Lopez-Fernandez, S, Garcia-Alija, M, Guerin, M.E. | | Deposit date: | 2016-05-31 | | Release date: | 2016-09-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structural Basis of Glycogen Biosynthesis Regulation in Bacteria.
Structure, 24, 2016
|
|
8BX5
 
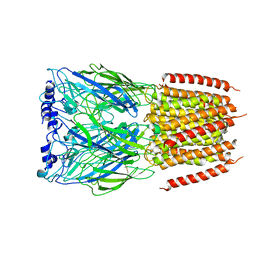 | |
8BXD
 
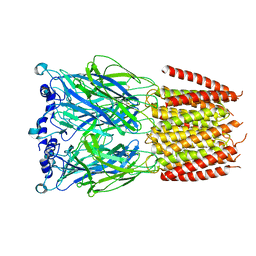 | |
4XMX
 
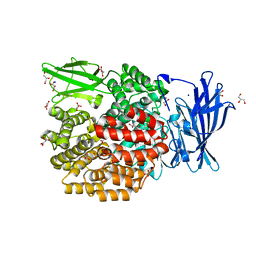 | |
4XN5
 
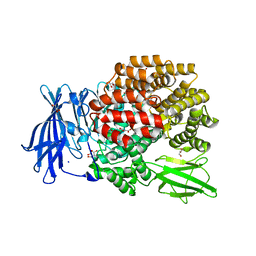 | |
8BXF
 
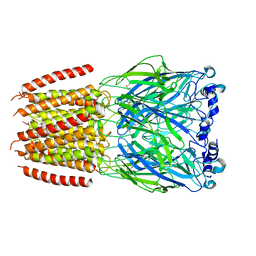 | |
8BXB
 
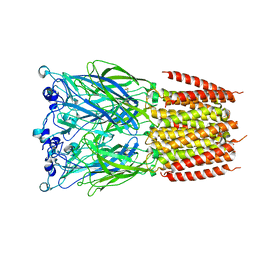 | |
8BXE
 
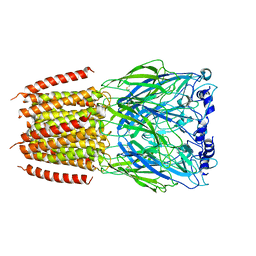 | |
4XQE
 
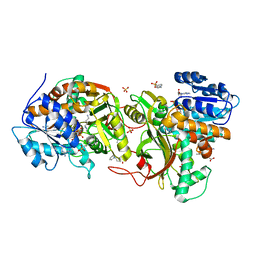 | |
5RYS
 
 | | EPB41L3 PanDDA analysis group deposition -- Crystal Structure of the FERM domain of human EPB41L3 in complex with Z1593306637 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Isoform 2 of Band 4.1-like protein 3, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | EPB41L3 PanDDA analysis group deposition
To Be Published
|
|
8BYI
 
 | | Alvinella pompejana nicotinic acetylcholine receptor Alpo4 in complex with CHAPS(Alpo4_CHAPS) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Acetylcholine receptor, ... | | Authors: | De Gieter, S, Efremov, R.G, Ulens, C. | | Deposit date: | 2022-12-13 | | Release date: | 2023-07-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Sterol derivative binding to the orthosteric site causes conformational changes in an invertebrate Cys-loop receptor.
Elife, 12, 2023
|
|
5RZ4
 
 | | EPB41L3 PanDDA analysis group deposition -- Crystal Structure of the FERM domain of human EPB41L3 in complex with Z2856434909 | | Descriptor: | (2R)-1-{[(2,6-dichlorophenyl)methyl]amino}propan-2-ol, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | EPB41L3 PanDDA analysis group deposition
To Be Published
|
|
5KU9
 
 | | Crystal structure of MCL1 with compound 1 | | Descriptor: | (3~{S})-3-azanyl-4-(4-bromophenyl)-~{N}-[(3~{S})-1-[2-[[(2~{R})-1-(3,4-dichlorophenyl)-4-(methylamino)-4-oxidanylidene-butan-2-yl]amino]-2-oxidanylidene-ethyl]-2-oxidanylidene-4,5-dihydro-3~{H}-1-benzazepin-3-yl]butanamide, Induced myeloid leukemia cell differentiation protein Mcl-1, SODIUM ION | | Authors: | Ferguson, A.D. | | Deposit date: | 2016-07-13 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure Based Design of Non-Natural Peptidic Macrocyclic Mcl-1 Inhibitors.
ACS Med Chem Lett, 8, 2017
|
|
5RZL
 
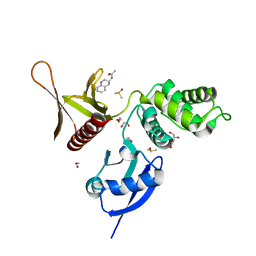 | | EPB41L3 PanDDA analysis group deposition -- Crystal Structure of the FERM domain of human EPB41L3 in complex with Z1492796719 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Isoform 2 of Band 4.1-like protein 3, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | EPB41L3 PanDDA analysis group deposition
To Be Published
|
|
1QTD
 
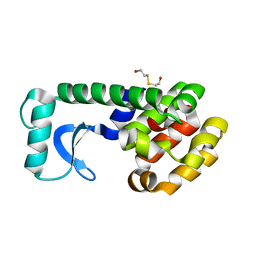 | |
5R1R
 
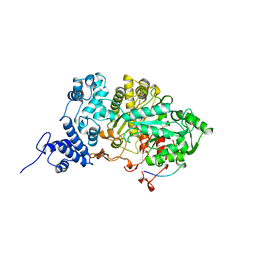 | | RIBONUCLEOTIDE REDUCTASE E441A MUTANT R1 PROTEIN FROM ESCHERICHIA COLI | | Descriptor: | RIBONUCLEOTIDE REDUCTASE R1 PROTEIN, RIBONUCLEOTIDE REDUCTASE R2 PROTEIN | | Authors: | Eriksson, M, Eklund, H. | | Deposit date: | 1997-09-17 | | Release date: | 1998-03-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A new mechanism-based radical intermediate in a mutant R1 protein affecting the catalytically essential Glu441 in Escherichia coli ribonucleotide reductase.
J.Biol.Chem., 272, 1997
|
|
7W57
 
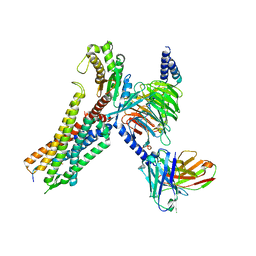 | | Cryo-EM structure of the neuromedin S-bound neuromedin U receptor 2-Gq protein complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(q) subunit alpha, ... | | Authors: | You, C, Xu, H.E, Jiang, Y. | | Deposit date: | 2021-11-29 | | Release date: | 2022-04-20 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into the peptide selectivity and activation of human neuromedin U receptors.
Nat Commun, 13, 2022
|
|
7W56
 
 | | Cryo-EM structure of the neuromedin S-bound neuromedin U receptor 1-Gq protein complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(q) subunit alpha, ... | | Authors: | You, C, Xu, H.E, Jiang, Y. | | Deposit date: | 2021-11-29 | | Release date: | 2022-04-20 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into the peptide selectivity and activation of human neuromedin U receptors.
Nat Commun, 13, 2022
|
|
1QAB
 
 | |
7W55
 
 | | Cryo-EM structure of the neuromedin U-bound neuromedin U receptor 2-Gq protein complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(q) subunit alpha, ... | | Authors: | You, C, Xu, H.E, Jiang, Y. | | Deposit date: | 2021-11-29 | | Release date: | 2022-04-20 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into the peptide selectivity and activation of human neuromedin U receptors.
Nat Commun, 13, 2022
|
|
7DD5
 
 | | Structure of Calcium-Sensing Receptor in complex with NPS-2143 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-chloro-6-[(2R)-2-hydroxy-3-{[2-methyl-1-(naphthalen-2-yl)propan-2-yl]amino}propoxy]benzonitrile, ... | | Authors: | Wen, T.L, Yang, X, Shen, Y.Q. | | Deposit date: | 2020-10-27 | | Release date: | 2021-06-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for activation and allosteric modulation of full-length calcium-sensing receptor.
Sci Adv, 7, 2021
|
|
7W53
 
 | | Cryo-EM structure of the neuromedin U-bound neuromedin U receptor 1-Gq protein complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(q) subunit alpha, ... | | Authors: | You, C, Xu, H.E, Jiang, Y. | | Deposit date: | 2021-11-29 | | Release date: | 2022-04-20 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into the peptide selectivity and activation of human neuromedin U receptors.
Nat Commun, 13, 2022
|
|
