1ERA
 
 | |
2MJC
 
 | | Zn-binding domain of eukaryotic translation initiation factor 3, subunit G | | Descriptor: | Eukaryotic translation initiation factor 3 subunit G, ZINC ION | | Authors: | Al-Abdul-Wahid, M, Menade, M, Xie, J, Kozlov, G, Gehring, K. | | Deposit date: | 2014-01-03 | | Release date: | 2015-01-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the Zn-binding domain of eukaryotic translation initiation factor 3, subunit G
To be Published
|
|
2CW6
 
 | | Crystal Structure of Human HMG-CoA Lyase: Insights into Catalysis and the Molecular Basis for Hydroxymethylglutaric Aciduria | | Descriptor: | 3-HYDROXYPENTANEDIOIC ACID, Hydroxymethylglutaryl-CoA lyase, mitochondrial, ... | | Authors: | Fu, Z, Runquist, J.A, Hunt, J.F, Miziorko, H.M, Kim, J.-J.P. | | Deposit date: | 2005-06-17 | | Release date: | 2005-12-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human 3-hydroxy-3-methylglutaryl-CoA Lyase: insights into catalysis and the molecular basis for hydroxymethylglutaric aciduria
J.Biol.Chem., 281, 2006
|
|
3WRR
 
 | |
3WRS
 
 | |
3WRQ
 
 | |
1ILO
 
 | | NMR structure of a thioredoxin, MtH895, from the archeon Methanobacterium thermoautotrophicum strain delta H. | | Descriptor: | conserved hypothetical protein MtH895 | | Authors: | Bhattacharyya, S, Habibi-Nazhad, B, Slupsky, C.M, Sykes, B.D, Wishart, D.S, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2001-05-08 | | Release date: | 2001-11-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Identification of a novel archaebacterial thioredoxin: determination of function through structure.
Biochemistry, 41, 2002
|
|
5T2P
 
 | | Hepatitis B virus core protein Y132A mutant in complex with sulfamoylbenzamide (SBA_R01) | | Descriptor: | 4-fluoranyl-3-(4-oxidanylpiperidin-1-yl)sulfonyl-~{N}-[3,4,5-tris(fluoranyl)phenyl]benzamide, CHLORIDE ION, Core protein, ... | | Authors: | Zhou, Z, Xu, Z.H. | | Deposit date: | 2016-08-24 | | Release date: | 2017-02-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.693 Å) | | Cite: | Heteroaryldihydropyrimidine (HAP) and Sulfamoylbenzamide (SBA) Inhibit Hepatitis B Virus Replication by Different Molecular Mechanisms.
Sci Rep, 7, 2017
|
|
3WRO
 
 | |
3WRN
 
 | |
2AEX
 
 | | The 1.58A Crystal Structure of Human Coproporphyrinogen Oxidase Reveals the Structural Basis of Hereditary Coproporphyria | | Descriptor: | CITRIC ACID, Coproporphyrinogen III oxidase, mitochondrial | | Authors: | Lee, D.S, Flachsova, E, Bodnarova, M, Demeler, B, Martasek, P, Raman, C.S. | | Deposit date: | 2005-07-24 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural basis of hereditary coproporphyria.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1PSB
 
 | | Solution structure of calcium loaded S100B complexed to a peptide from N-Terminal regulatory domain of NDR kinase. | | Descriptor: | Ndr Ser/Thr kinase-like protein, S-100 protein, beta chain | | Authors: | Bhattacharya, S, Large, E, Heizmann, C.W, Hemmings, B, Chazin, W.J. | | Deposit date: | 2003-06-21 | | Release date: | 2003-12-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the Ca(2+)/S100B/NDR Kinase Peptide Complex: Insights into S100 Target Specificity and Activation of the Kinase.
Biochemistry, 42, 2003
|
|
1Q0P
 
 | | A domain of Factor B | | Descriptor: | Complement factor B, MANGANESE (II) ION | | Authors: | Bhattacharya, A.A, Liddington, R.C. | | Deposit date: | 2003-07-17 | | Release date: | 2004-03-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the A Domain from Complement Factor B Reveals an Integrin-like Open Conformation.
STRUCTURE, 12, 2004
|
|
1LSJ
 
 | |
1M75
 
 | |
1E7E
 
 | |
1M76
 
 | |
1PYT
 
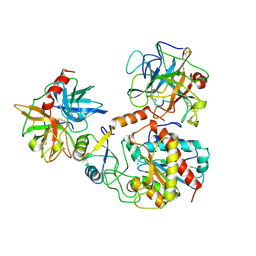 | | TERNARY COMPLEX OF PROCARBOXYPEPTIDASE A, PROPROTEINASE E, AND CHYMOTRYPSINOGEN C | | Descriptor: | CALCIUM ION, CHYMOTRYPSINOGEN C, PROCARBOXYPEPTIDASE A, ... | | Authors: | Gomis-Ruth, F.X, Gomez, M, Bode, W, Huber, R, Aviles, F.X. | | Deposit date: | 1995-06-21 | | Release date: | 1997-01-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The three-dimensional structure of the native ternary complex of bovine pancreatic procarboxypeptidase A with proproteinase E and chymotrypsinogen C.
EMBO J., 14, 1995
|
|
1E7I
 
 | |
2KJD
 
 | |
1E7F
 
 | |
1YSM
 
 | | NMR Structure of N-terminal domain (Residues 1-77) of Siah-Interacting Protein. | | Descriptor: | Calcyclin-binding protein | | Authors: | Bhattacharya, S, Lee, Y.T, Michowski, W, Jastrzebska, B, Filipek, A, Kuznicki, J, Chazin, W.J. | | Deposit date: | 2005-02-08 | | Release date: | 2005-07-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The Modular Structure of SIP Facilitates Its Role in Stabilizing Multiprotein Assemblies.
Biochemistry, 44, 2005
|
|
1E7H
 
 | |
1E7G
 
 | |
2M0U
 
 | | Complex structure of C-terminal CFTR peptide and extended PDZ1 domain from NHERF1 | | Descriptor: | C-terminal CFTR peptide, Na(+)/H(+) exchange regulatory cofactor NHE-RF1 | | Authors: | Bhattacharya, S, Ju, J.H, Cowburn, D, Bu, Z. | | Deposit date: | 2012-11-06 | | Release date: | 2013-04-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ligand-Induced Dynamic Changes in Extended PDZ Domains from NHERF1.
J.Mol.Biol., 425, 2013
|
|
