6YI9
 
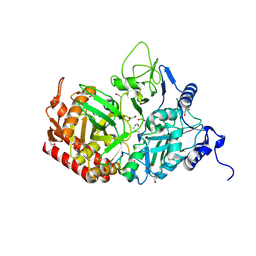 | | Crystal structure of the rat cytosolic PCK1, acetylated on Lys244 | | Descriptor: | 1,2-ETHANEDIOL, Phosphoenolpyruvate carboxykinase, cytosolic [GTP] | | Authors: | Latorre-Muro, P, Baeza, J, Hurtado-Guerrero, R, Hicks, T, Delso, I, Hernandez-Ruiz, C, Velazquez-Campoy, A, Lawton, A.J, Angulo, J, Denu, J.M, Carrodeguas, J.A. | | Deposit date: | 2020-04-01 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Self-acetylation at the active site of phosphoenolpyruvate carboxykinase (PCK1) controls enzyme activity.
J.Biol.Chem., 296, 2021
|
|
6QS1
 
 | |
4R43
 
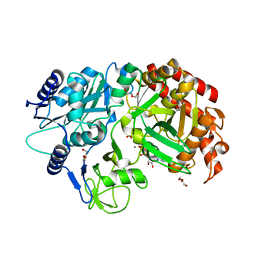 | | Crystal Structure Analysis of MTB PEPCK | | Descriptor: | DI(HYDROXYETHYL)ETHER, GUANOSINE-5'-DIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Brynda, J, Dostal, J, Pichova, I, Snasel, J, Fanfrlik, J, Machova, I. | | Deposit date: | 2014-08-19 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Functional Studies of Phosphoenolpyruvate Carboxykinase from Mycobacterium tuberculosis.
Plos One, 10, 2015
|
|
4GMZ
 
 | |
4GNP
 
 | |
4GNO
 
 | |
4GNL
 
 | |
4GMM
 
 | |
4GMW
 
 | |
4GMU
 
 | |
4GNQ
 
 | |
4GNM
 
 | |
6E59
 
 | | Crystal structure of the human NK1 tachykinin receptor | | Descriptor: | 1-(4-{[(2R,3S)-2-{(1R)-1-[3,5-bis(trifluoromethyl)phenyl]ethoxy}-3-(4-fluorophenyl)morpholin-4-yl]methyl}-1H-1,2,3-triazol-5-yl)-N,N-dimethylmethanamine, Substance-P receptor, GlgA glycogen synthase, ... | | Authors: | Yin, J, Clark, L, Chapman, K, Shao, Z, Borek, D, Xu, Q, Wang, J, Rosenbaum, D.M. | | Deposit date: | 2018-07-19 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of the human NK1tachykinin receptor.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6XU4
 
 | | Crystal structure of the genetically-encoded FGCaMP calcium indicator in its calcium-bound state | | Descriptor: | CALCIUM ION, FGCamp | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Korzhenevskiy, D.A, Barykina, N.V, Subach, O.M, Subach, F.V. | | Deposit date: | 2020-01-17 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | FGCaMP7, an Improved Version of Fungi-Based Ratiometric Calcium Indicator for In Vivo Visualization of Neuronal Activity.
Int J Mol Sci, 21, 2020
|
|
6ET6
 
 | | Crystal structure of muramidase from Acinetobacter baumannii AB 5075UW prophage | | Descriptor: | GLYCEROL, Lysozyme, SULFATE ION | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Sykilinda, N.N, Shneider, M.M, Miroshnikov, K.A, Popov, V.O. | | Deposit date: | 2017-10-25 | | Release date: | 2018-09-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of anAcinetobacterBroad-Range Prophage Endolysin Reveals a C-Terminal alpha-Helix with the Proposed Role in Activity against Live Bacterial Cells.
Viruses, 10, 2018
|
|
8VJX
 
 | |
3CWW
 
 | | Crystal Structure of IDE-bradykinin complex | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, ACETATE ION, Insulin-degrading enzyme, ... | | Authors: | Malito, E, Tang, W.J. | | Deposit date: | 2008-04-23 | | Release date: | 2008-11-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Molecular Bases for the Recognition of Short Peptide Substrates and Cysteine-Directed Modifications of Human Insulin-Degrading Enzyme
Biochemistry, 47, 2008
|
|
2GMV
 
 | | PEPCK complex with a GTP-competitive inhibitor | | Descriptor: | MANGANESE (II) ION, N-(4-{[3-BUTYL-1-(2-FLUOROBENZYL)-2,6-DIOXO-2,3,6,7-TETRAHYDRO-1H-PURIN-8-YL]METHYL}PHENYL)-1-METHYL-1H-IMIDAZOLE-4-SULFONAMIDE, PHOSPHOENOLPYRUVATE, ... | | Authors: | Dunten, P. | | Deposit date: | 2006-04-07 | | Release date: | 2007-05-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | C-8 Modifications of 3-alkyl-1,8-dibenzylxanthines as inhibitors of human cytosolic phosphoenolpyruvate carboxykinase.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
4ASR
 
 | | Crystal structure of ANCE in complex with Thr6-Bradykinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANGIOTENSIN-CONVERTING ENZYME, BRADYKININ, ... | | Authors: | Akif, M, Masuyer, G, Sturrock, E.D, Isaac, R.E, Acharya, K.R. | | Deposit date: | 2012-05-02 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of Peptide Recognition by the Angiotensin-I Converting Enzyme Homologue Ance from Drosophila Melanogaster
FEBS J., 279, 2012
|
|
4ASQ
 
 | | Crystal structure of ANCE in complex with Bradykinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANGIOTENSIN-CONVERTING ENZYME, BRADYKININ, ... | | Authors: | Akif, M, Masuyer, G, Sturrock, E.D, Isaac, R.E, Acharya, K.R. | | Deposit date: | 2012-05-02 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural Basis of Peptide Recognition by the Angiotensin-I Converting Enzyme Homologue Ance from Drosophila Melanogaster
FEBS J., 279, 2012
|
|
1VZK
 
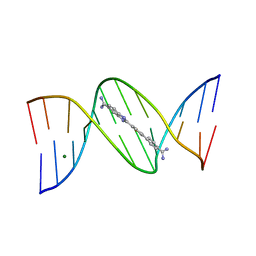 | | A Thiophene Based Diamidine Forms a "Super" AT Binding Minor Groove Agent | | Descriptor: | 2-(5-{4-[AMINO(IMINO)METHYL]PHENYL}-2-THIENYL)-1H-BENZIMIDAZOLE-6- CARBOXIMIDAMIDE DIHYDROCHLORIDE, 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP *CP*G)-3', MAGNESIUM ION | | Authors: | Mallena, S, Lee, M.P.H, Bailly, C, Neidle, S, Kumar, A, Boykin, D.W, Wilson, W.D. | | Deposit date: | 2004-05-20 | | Release date: | 2004-10-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Thiophene-Based Diamidine Forms a "Super" at Binding Minor Groove Agent
J.Am.Chem.Soc., 142, 2004
|
|
4AA2
 
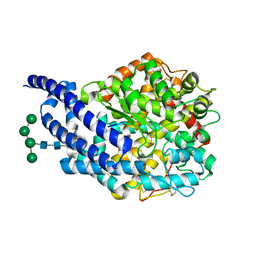 | | Crystal structure of ANCE in complex with bradykinin potentiating peptide b | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANGIOTENSIN-CONVERTING ENZYME, BRADYKININ-POTENTIATING PEPTIDE B, ... | | Authors: | Isaac, R.E, Akif, M, Schwager, S.L.U, Masuyer, G, Sturrock, E.D, Acharya, K.R. | | Deposit date: | 2011-11-30 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural Basis of Peptide Recognition by the Angiotensin-I Converting Enzyme Homologue Ance from Drosophila Melanogaster
FEBS J., 279, 2012
|
|
2GFR
 
 | |
2MCE
 
 | |
2OLR
 
 | | Crystal structure of Escherichia coli phosphoenolpyruvate carboxykinase complexed with carbon dioxide, Mg2+, ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CARBON DIOXIDE, CHLORIDE ION, ... | | Authors: | Cotelesage, J.J, Delbaere, L.T, Goldie, H, Puttick, J, Rajabi, B, Novakovski, B. | | Deposit date: | 2007-01-19 | | Release date: | 2007-06-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | How does an enzyme recognize CO2?
Int.J.Biochem.Cell Biol., 39, 2007
|
|
