2F1W
 
 | | Crystal structure of the TRAF-like domain of HAUSP/USP7 | | Descriptor: | CALCIUM ION, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Hu, M, Gu, L, Jeffrey, P.D, Shi, Y. | | Deposit date: | 2005-11-15 | | Release date: | 2006-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Basis of Competitive Recognition of p53 and MDM2 by HAUSP/USP7: Implications for the Regulation of the p53-MDM2 Pathway.
Plos Biol., 4, 2006
|
|
8D4Z
 
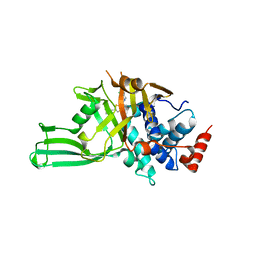 | | Crystal structure of USP7 in complex with allosteric inhibitor FX1-3763 | | Descriptor: | 1-({(7M)-7-[1-(azetidin-3-yl)-6-chloro-1,2,3,4-tetrahydroquinolin-8-yl]thieno[3,2-b]pyridin-2-yl}methyl)pyrrolidine-2,5-dione, GLYCEROL, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Bell, J.A. | | Deposit date: | 2022-06-03 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Novel USP7 inhibitors demonstrate potent anti-cancer activity in models of AML, synergy with BCL2 inhibition, and a differentiated mechanism of action
To Be Published
|
|
7CM2
 
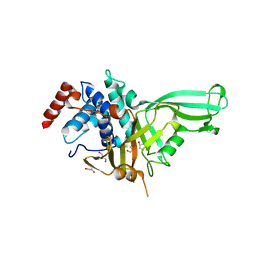 | | The Crystal Structure of human USP7 USP domain from Biortus | | Descriptor: | GLYCEROL, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Wang, F, Cheng, W, Lv, Z, Lin, D, Zhu, B, Miao, Q, Bao, X, Shang, H. | | Deposit date: | 2020-07-24 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Crystal Structure of human USP7 USP domain from Biortus.
To Be Published
|
|
3MQS
 
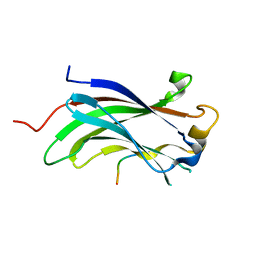 | |
4PYZ
 
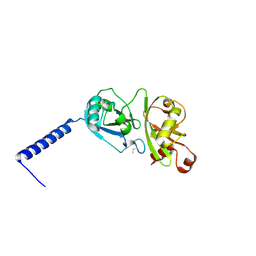 | | Crystal structure of the first two Ubl domains of Deubiquitylase USP7 | | Descriptor: | UNKNOWN ATOM OR ION, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Walker, J.R, Dong, A, Ong, M.S, Dhe-Paganon, S, Kania, J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-03-28 | | Release date: | 2014-04-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Crystal structure of the first two Ubl domains of Deubiquitylase USP7
to be published
|
|
2KVR
 
 | | Solution NMR structure of human ubiquitin specific protease Usp7 UBL domain (residues 537-664). NESG target hr4395c/ SGC-Toronto | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Bezsonova, I, Lemak, A, Avvakumov, G, Xue, S, Dhe-Paganon, S, Montelione, G.T, Arrowsmith, C, Northeast Structural Genomics Consortium (NESG), Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-25 | | Release date: | 2010-04-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | solution structure of the ubiquitin specific protease Usp7 ubiquitin-like domain
To be Published
|
|
3MQR
 
 | |
5C56
 
 | | Crystal structure of USP7/HAUSP in complex with ICP0 | | Descriptor: | Ubiquitin E3 ligase ICP0, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Cheng, J, Li, Z, Gong, R, Fang, J, Yang, Y, Sun, C, Yang, H, Xu, Y. | | Deposit date: | 2015-06-19 | | Release date: | 2015-07-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.685 Å) | | Cite: | Molecular mechanism for the substrate recognition of USP7.
Protein Cell, 6, 2015
|
|
4Z96
 
 | | Crystal structure of DNMT1 in complex with USP7 | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Zhang, Z.M, Song, J. | | Deposit date: | 2015-04-09 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of DNMT1 in complex with USP7
To Be Published
|
|
7XHK
 
 | | High-resolution X-ray cocrystal structure of USP7 in complex with LX04-46 | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 7, ~{N}-[[4-[6-[[1-[[2-chloranyl-4-(furan-2-yl)phenyl]methyl]-4-oxidanyl-piperidin-4-yl]methyl]-2-methyl-7-oxidanylidene-pyrazolo[4,3-d]pyrimidin-3-yl]phenyl]methyl]methanamide | | Authors: | Sun, H.B, Wen, X.A. | | Deposit date: | 2022-04-08 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | High-resolution X-ray cocrystal structure of USP7 in complex with LX04-46
To Be Published
|
|
7XHH
 
 | | High-resolution X-ray cocrystal structure of USP7 in complex with X4 | | Descriptor: | 3-[4-(aminomethyl)phenyl]-6-[[1-[[2-chloranyl-4-(1,2,4-oxadiazol-3-yl)phenyl]methyl]-4-oxidanyl-piperidin-4-yl]methyl]-2-methyl-pyrazolo[4,3-d]pyrimidin-7-one, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Sun, H.B, Wen, X.A. | | Deposit date: | 2022-04-08 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-resolution X-ray cocrystal structure of USP7 in complex with X4
To Be Published
|
|
4YSI
 
 | | Structure of USP7 with a novel viral protein | | Descriptor: | GLYCEROL, SER-PRO-GLY-GLU-GLY-PRO-SER-GLY, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Chavoshi, S, Saridakis, V. | | Deposit date: | 2015-03-17 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Structure of USP7 with a novel viral protein
J.Biol.Chem., 2016
|
|
4Z97
 
 | |
5VSB
 
 | | Structure of DUB complex | | Descriptor: | 7-chloro-3-{[4-hydroxy-1-(3-phenylpropanoyl)piperidin-4-yl]methyl}quinazolin-4(3H)-one, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2017-05-11 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Guided Development of a Potent and Selective Non-covalent Active-Site Inhibitor of USP7.
Cell Chem Biol, 24, 2017
|
|
5VS6
 
 | | Structure of DUB complex | | Descriptor: | ACETATE ION, GLYCEROL, N-[3-({4-hydroxy-1-[(3R)-3-phenylbutanoyl]piperidin-4-yl}methyl)-4-oxo-3,4-dihydroquinazolin-7-yl]-3-(4-methylpiperazin-1-yl)propanamide, ... | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2017-05-11 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structure-Guided Development of a Potent and Selective Non-covalent Active-Site Inhibitor of USP7.
Cell Chem Biol, 24, 2017
|
|
5VSK
 
 | | Structure of DUB complex | | Descriptor: | 7-chloro-3-({4-hydroxy-1-[(3S)-3-phenylbutanoyl]piperidin-4-yl}methyl)quinazolin-4(3H)-one, Ubiquitin carboxyl-terminal hydrolase 7, ZINC ION | | Authors: | Seo, H.-Y, Dhe-Paganon, S. | | Deposit date: | 2017-05-11 | | Release date: | 2017-12-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | Structure-Guided Development of a Potent and Selective Non-covalent Active-Site Inhibitor of USP7.
Cell Chem Biol, 24, 2017
|
|
2F1Z
 
 | | Crystal structure of HAUSP | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Hu, M, Gu, L, Jeffrey, P.D, Shi, Y. | | Deposit date: | 2005-11-15 | | Release date: | 2006-02-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis of Competitive Recognition of p53 and MDM2 by HAUSP/USP7: Implications for the Regulation of the p53-MDM2 Pathway.
Plos Biol., 4, 2006
|
|
5VBT
 
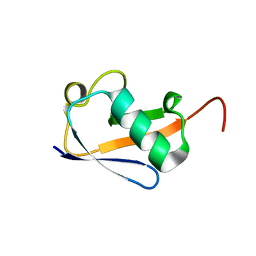 | | Crystal structure of a highly specific and potent USP7 ubiquitin variant inhibitor | | Descriptor: | UBH04 | | Authors: | DONG, A, DONG, X, LIU, L, GUO, Y, LI, Y, ZHANG, W, WALKER, J.R, SIDHU, S, Bountra, C, Arrowsmith, C.H, Edwards, A.M, TONG, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-03-30 | | Release date: | 2017-06-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystal structure of a highly specific and potent USP7 ubiquitin variant inhibitor
to be published
|
|
9DEO
 
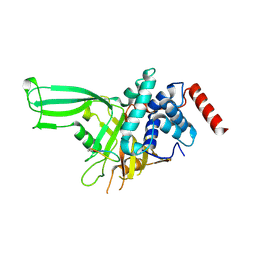 | | USP7 in complex with macrocycle inhibitor MC08 | | Descriptor: | Macrocycle peptide MC08, SULFATE ION, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Ultsch, M, Tenorio, C.A, Dueber, E.C, Harris, S.F. | | Deposit date: | 2024-08-29 | | Release date: | 2025-03-05 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery and characterization of potent macrocycle inhibitors of ubiquitin-specific protease-7.
Structure, 33, 2025
|
|
9DEL
 
 | | USP7 in complex with macrocycle MC03 | | Descriptor: | Macrocycle peptide MC03, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Ultsch, M, Tenorio, C.A, Dueber, E.C, Harris, S.F. | | Deposit date: | 2024-08-29 | | Release date: | 2025-03-05 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery and characterization of potent macrocycle inhibitors of ubiquitin-specific protease-7.
Structure, 33, 2025
|
|
9DEK
 
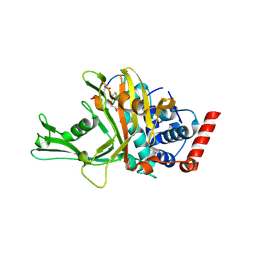 | | USP7 in complex with macrocycle inhibitor MC02 | | Descriptor: | GLYCEROL, Macrocycle peptide MC02, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Rouge, L, Ultsch, M, Holden, J.K, Dueber, E.C, Harris, S.F. | | Deposit date: | 2024-08-29 | | Release date: | 2025-03-05 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and characterization of potent macrocycle inhibitors of ubiquitin-specific protease-7.
Structure, 33, 2025
|
|
9DEN
 
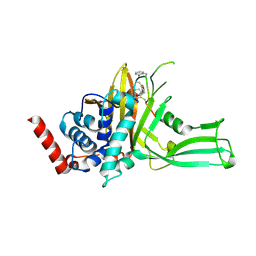 | | USP7 in complex with macrocycle MC07 | | Descriptor: | ETHANOL, Macrocycle peptide MC07, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Ultsch, M, Tenorio, C.A, Dueber, E.C, Harris, S.F. | | Deposit date: | 2024-08-29 | | Release date: | 2025-03-05 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Discovery and characterization of potent macrocycle inhibitors of ubiquitin-specific protease-7.
Structure, 33, 2025
|
|
9DEM
 
 | | USP7 in complex with macrocycle MC04 | | Descriptor: | Macrocycle peptide MC04, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Rouge, L, Ultsch, M, Dueber, E.C, Harris, S.F. | | Deposit date: | 2024-08-29 | | Release date: | 2025-03-05 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Discovery and characterization of potent macrocycle inhibitors of ubiquitin-specific protease-7.
Structure, 33, 2025
|
|
9DEP
 
 | | USP7 in complex with macrocycle MC09 | | Descriptor: | Macrocycle peptide MC09, SULFATE ION, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Ultsch, M, Tenorio, C.A, Dueber, E.C, Harris, S.F. | | Deposit date: | 2024-08-29 | | Release date: | 2025-03-05 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Discovery and characterization of potent macrocycle inhibitors of ubiquitin-specific protease-7.
Structure, 33, 2025
|
|
4JJQ
 
 | | Crystal structure of usp7-ntd with an e2 enzyme | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 7, Ubiquitin-conjugating enzyme E2 E1 | | Authors: | Saridakis, V. | | Deposit date: | 2013-03-08 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Ubiquitin-specific protease 7 is a regulator of ubiquitin-conjugating enzyme UbE2E1.
J. Biol. Chem., 288, 2013
|
|
