4XHV
 
 | | Crystal structure of Drosophila Spinophilin-PDZ and a C-terminal peptide of Neurexin | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, LP20995p, ... | | Authors: | Driller, J.H, Muhammad, K.G.H, Reddy, S, Rey, U, Boehme, M.A, Hollmann, C, Ramesh, N, Depner, H, Luetzkendorf, J, Matkovic, T, Bergeron, D, Quentin, C, Schmoranzer, J, Goettfert, F, Holt, M, Wahl, M.C, Hell, S.W, Walter, A, Sigrist, S.J, Loll, B. | | Deposit date: | 2015-01-06 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Presynaptic spinophilin tunes neurexin signalling to control active zone architecture and function.
Nat Commun, 6, 2015
|
|
4XNE
 
 | | Crystal structure of ERK2 in complex with an inhibitor | | Descriptor: | 1-phenyl-1H-1,2,4-triazole-3,5-diamine, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Gelin, M, Allemand, F, Labesse, G, Guichou, J.F. | | Deposit date: | 2015-01-15 | | Release date: | 2015-08-12 | | Last modified: | 2015-08-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Combining `dry' co-crystallization and in situ diffraction to facilitate ligand screening by X-ray crystallography.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4U03
 
 | |
4U0M
 
 | |
6SM0
 
 | | Venus 66 p-Azido-L-Phenylalanin (azF) variant, dark grown | | Descriptor: | Green fluorescent protein, OXYGEN MOLECULE, SULFATE ION, ... | | Authors: | Rizkallah, P.J, Al Maslookhi, H.S, Jones, D.D. | | Deposit date: | 2019-08-21 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Stalling chromophore synthesis of the fluorescent protein Venus reveals the molecular basis of the final oxidation step.
Chem Sci, 12, 2021
|
|
4XLQ
 
 | |
4XP2
 
 | | Crystal structure of ERK2 in complex with an inhibitor | | Descriptor: | 1-phenyl-1H-1,2,4-triazole-3,5-diamine, DIMETHYL SULFOXIDE, Mitogen-activated protein kinase 1, ... | | Authors: | Gelin, M, Allemand, F, Labesse, G, Guichou, J.F. | | Deposit date: | 2015-01-16 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.748 Å) | | Cite: | Combining `dry' co-crystallization and in situ diffraction to facilitate ligand screening by X-ray crystallography.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6Q7S
 
 | | ERK2 mini-fragment binding | | Descriptor: | Mitogen-activated protein kinase 1, PHENOL, SULFATE ION | | Authors: | O'Reilly, M, Cleasby, A, Davies, T.G, Hall, R, Ludlow, F, Murray, C.W, Tisi, D, Jhoti, H. | | Deposit date: | 2018-12-13 | | Release date: | 2019-06-26 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design.
Drug Discov Today, 24, 2019
|
|
6SHX
 
 | | DNA mismatch repair proteins MLH1 and MLH3 | | Descriptor: | DNA mismatch repair protein MLH1, DNA mismatch repair protein MLH3, ZINC ION | | Authors: | Dai, J, Chervy, P, Legrand, P, Ropars, V, Charbonnier, J.B. | | Deposit date: | 2019-08-08 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis of the dual role of the Mlh1-Mlh3 endonuclease in MMR and in meiotic crossover formation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6SNS
 
 | | DNA mismatch repair proteins MLH1 and MLH3 | | Descriptor: | DNA mismatch repair protein MLH1, DNA mismatch repair protein MLH3, ZINC ION | | Authors: | Dai, J, Chervy, P, Legrand, P, Ropars, V, Charbonnier, J.B. | | Deposit date: | 2019-08-27 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular basis of the dual role of the Mlh1-Mlh3 endonuclease in MMR and in meiotic crossover formation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6SNV
 
 | | DNA mismatch repair proteins MLH1 and MLH3 | | Descriptor: | DNA mismatch repair protein MLH1, DNA mismatch repair protein MLH3, ZINC ION | | Authors: | Dai, J, Chervy, P, Legrand, P, Ropars, V, Charbonnier, J.B. | | Deposit date: | 2019-08-27 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular basis of the dual role of the Mlh1-Mlh3 endonuclease in MMR and in meiotic crossover formation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
2NWM
 
 | |
4Y1M
 
 | |
6Q7T
 
 | | ERK2 mini-fragment binding | | Descriptor: | 1,2-oxazol-3-amine, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | O'Reilly, M, Cleasby, A, Davies, T.G, Hall, R, Ludlow, F, Murray, C.W, Tisi, D, Jhoti, H. | | Deposit date: | 2018-12-13 | | Release date: | 2019-06-26 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design.
Drug Discov Today, 24, 2019
|
|
6TD2
 
 | |
6QAH
 
 | | ERK2 mini-fragment binding | | Descriptor: | 1-azanylpropylideneazanium, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | O'Reilly, M, Cleasby, A, Davies, T.G, Hall, R, Ludlow, F, Murray, C.W, Tisi, D, Jhoti, H. | | Deposit date: | 2018-12-19 | | Release date: | 2019-06-26 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design.
Drug Discov Today, 24, 2019
|
|
6T58
 
 | | Structure determination of the transactivation domain of p53 in complex with S100A4 using annexin A2 as a crystallization chaperone | | Descriptor: | CALCIUM ION, Cellular tumor antigen p53,Protein S100-A4,Protein S100-A4,Annexin A2, GLYCEROL | | Authors: | Ecsedi, P, Gogl, G, Nyitray, L. | | Deposit date: | 2019-10-15 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure Determination of the Transactivation Domain of p53 in Complex with S100A4 Using Annexin A2 as a Crystallization Chaperone.
Structure, 28, 2020
|
|
6R1T
 
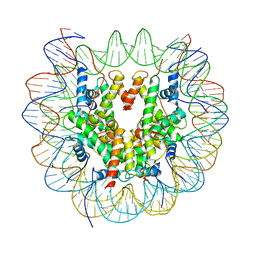 | | Structure of LSD2/NPAC-linker/nucleosome core particle complex: Class 1, free nuclesome | | Descriptor: | DNA (147-MER), HISTONE H2A, Histone H2A, ... | | Authors: | Marabelli, C, Pilotto, S, Chittori, S, Subramaniam, S, Mattevi, A. | | Deposit date: | 2019-03-15 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.02 Å) | | Cite: | A Tail-Based Mechanism Drives Nucleosome Demethylation by the LSD2/NPAC Multimeric Complex.
Cell Rep, 27, 2019
|
|
6R1U
 
 | | Structure of LSD2/NPAC-linker/nucleosome core particle complex: Class 2 | | Descriptor: | DNA (147-MER), FLAVIN-ADENINE DINUCLEOTIDE, Histone H2A, ... | | Authors: | Marabelli, C, Pilotto, S, Chittori, S, Subramaniam, S, Mattevi, A. | | Deposit date: | 2019-03-15 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.36 Å) | | Cite: | A Tail-Based Mechanism Drives Nucleosome Demethylation by the LSD2/NPAC Multimeric Complex.
Cell Rep, 27, 2019
|
|
6Q9A
 
 | |
6R25
 
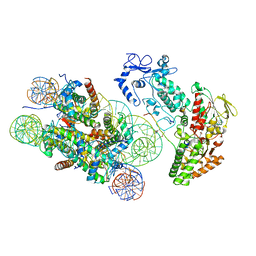 | | Structure of LSD2/NPAC-linker/nucleosome core particle complex: Class 3 | | Descriptor: | DNA (147-MER), FLAVIN-ADENINE DINUCLEOTIDE, H2B, ... | | Authors: | Marabelli, C, Pilotto, S, Chittori, S, Subramaniam, S, Mattevi, A. | | Deposit date: | 2019-03-15 | | Release date: | 2019-04-24 | | Method: | ELECTRON MICROSCOPY (4.61 Å) | | Cite: | A Tail-Based Mechanism Drives Nucleosome Demethylation by the LSD2/NPAC Multimeric Complex.
Cell Rep, 27, 2019
|
|
6OUO
 
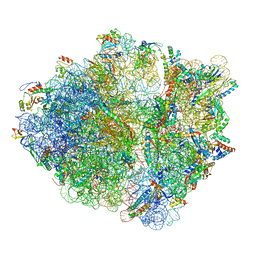 | | RF2 accommodated state bound 70S complex at long incubation time | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fu, Z, Indrisiunaite, G, Kaledhonkar, S, Shah, B, Sun, M, Chen, B, Grassucci, R.A, Ehrenberg, M, Frank, J. | | Deposit date: | 2019-05-05 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy.
Nat Commun, 10, 2019
|
|
4XJ0
 
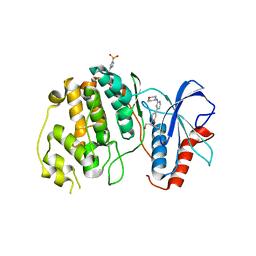 | | Crystal structure of ERK2 in complex with an inhibitor 14K | | Descriptor: | 1-[(1S)-1-(4-chloro-3-fluorophenyl)-2-hydroxyethyl]-4-[2-(tetrahydro-2H-pyran-4-ylamino)pyrimidin-4-yl]pyridin-2(1H)-one, Mitogen-activated protein kinase 1 | | Authors: | Yin, J, Wang, W. | | Deposit date: | 2015-01-08 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Discovery of highly potent, selective, and efficacious small molecule inhibitors of ERK1/2.
J.Med.Chem., 58, 2015
|
|
6RCU
 
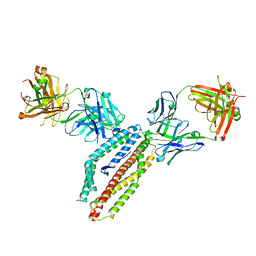 | | PfRH5 bound to monoclonal antibodies R5.004 and R5.016 | | Descriptor: | R5.004 heavy chain, R5.004 light chain, R5.016 heavy chain, ... | | Authors: | Alanine, D.W.G, Draper, S.J, Higgins, M.K. | | Deposit date: | 2019-04-11 | | Release date: | 2019-06-26 | | Last modified: | 2019-08-21 | | Method: | X-RAY DIFFRACTION (4.005 Å) | | Cite: | Human Antibodies that Slow Erythrocyte Invasion Potentiate Malaria-Neutralizing Antibodies.
Cell, 178, 2019
|
|
6OFJ
 
 | |
