9GI4
 
 | | TFIIIC5 DNA binding domain | | Descriptor: | General transcription factor 3C polypeptide 5, PHOSPHATE ION | | Authors: | Leen, E, Bayliss, R. | | Deposit date: | 2024-08-16 | | Release date: | 2025-08-27 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | N-myc interacts with TFIIIC via the DNA binding interface of TFIIIC5.
To Be Published
|
|
2PAB
 
 | |
2PAF
 
 | |
1MG8
 
 | | NMR structure of ubiquitin-like domain in murine Parkin | | Descriptor: | Parkin | | Authors: | Tashiro, M, Okubo, S, Shimotakahara, S, Hatanaka, H, Yasuda, H, Kainosho, M, Yokoyama, S, Shindo, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-08-15 | | Release date: | 2003-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of ubiquitin-like domain in PARKIN: Gene product of familial Parkinson's disease.
J.Biomol.NMR, 25, 2003
|
|
1MGX
 
 | | COAGULATION FACTOR, MG(II), NMR, 7 STRUCTURES (BACKBONE ATOMS ONLY) | | Descriptor: | COAGULATION FACTOR IX | | Authors: | Freedman, S.J, Furie, B.C, Furie, B, Baleja, J.D. | | Deposit date: | 1995-06-21 | | Release date: | 1996-11-08 | | Last modified: | 2025-03-26 | | Method: | SOLUTION NMR | | Cite: | Identification of the phospholipid binding site in the vitamin K-dependent blood coagulation protein factor IX.
J.Biol.Chem., 271, 1996
|
|
7SEX
 
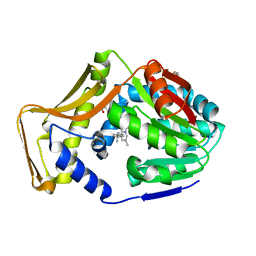 | | M. tb EgtD in complex with TGX221 | | Descriptor: | 7-methyl-2-morpholin-4-yl-9-[(1~{R})-1-phenylazanylethyl]-3~{H}-pyrido[1,2-a]pyrimidin-4-one, GLYCEROL, Histidine N-alpha-methyltransferase | | Authors: | Sudasinghe, T.D, Ronning, D.R. | | Deposit date: | 2021-10-02 | | Release date: | 2021-12-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inhibitors of Mycobacterium tuberculosis EgtD target both substrate binding sites to limit hercynine production.
Sci Rep, 11, 2021
|
|
1MMG
 
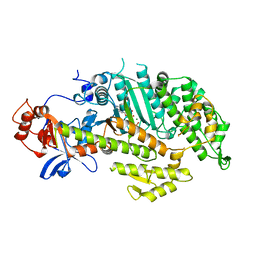 | | X-RAY STRUCTURES OF THE MGADP, MGATPGAMMAS, AND MGAMPPNP COMPLEXES OF THE DICTYOSTELIUM DISCOIDEUM MYOSIN MOTOR DOMAIN | | Descriptor: | MAGNESIUM ION, MYOSIN, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Gulick, A.M, Bauer, C.B, Thoden, J.B, Rayment, I. | | Deposit date: | 1997-07-18 | | Release date: | 1997-12-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray structures of the MgADP, MgATPgammaS, and MgAMPPNP complexes of the Dictyostelium discoideum myosin motor domain.
Biochemistry, 36, 1997
|
|
9GI5
 
 | | Fe(II)-2-oxoglutarate-dependent Oryza sativa dioxygenase HSL1 in complex with 2-oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Dhingra, S, Alshref, F.M, Farcas, I, Allen, M.D, Brewitz, L, Schofield, C.J. | | Deposit date: | 2024-08-16 | | Release date: | 2025-08-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Fe(II)-2-oxoglutarate-dependent Oryza sativa dioxygenase HSL1 in complex with 2-oxoglutarate
To Be Published
|
|
9GA0
 
 | |
9GF6
 
 | |
1MN8
 
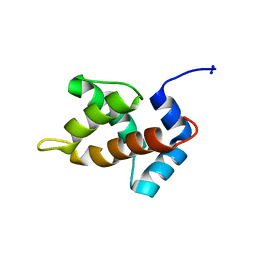 | | Structure of Moloney Murine Leukaemia Virus Matrix Protein | | Descriptor: | Core protein p15 | | Authors: | Riffel, N, Harlos, K, Iourin, O, Rao, Z, Kingsman, A, Stuart, D, Fry, E. | | Deposit date: | 2002-09-05 | | Release date: | 2003-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Atomic resolution structure of Moloney murine leukaemia virus matrix protein and its relationship to other retroviral matrix proteins.
Structure, 10, 2002
|
|
1MH5
 
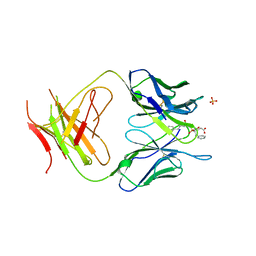 | | The Structure Of The Complex Of The Fab Fragment Of The Esterolytic Antibody MS6-164 and A Transition-State Analog | | Descriptor: | IMMUNOGLOBULIN MS6-164, N-{[2-({[1-(4-CARBOXYBUTANOYL)AMINO]-2-PHENYLETHYL}-HYDROXYPHOSPHINYL)OXY]ACETYL}-2-PHENYLETHYLAMINE, SULFATE ION | | Authors: | Ruzheinikov, S.N, Muranova, T.A, Sedelnikova, S.E, Partridge, L.J, Blackburn, G.M, Murray, I.A, Kakinuma, H, Takashi, N, Shimazaki, K, Sun, J, Nishi, Y, Rice, D.W. | | Deposit date: | 2002-08-19 | | Release date: | 2003-09-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-resolution crystal structure of the Fab-fragments of a family of mouse catalytic antibodies with esterase activity
J.Mol.Biol., 332, 2003
|
|
9GF5
 
 | | CRYSTAL STRUCTURE OF COMPLEX OF ASO BINDING FAB FRAGMENT IN COMPLEX WITH ASO980 | | Descriptor: | 1,2-ETHANEDIOL, 1-[(2R,4S,5R)-4-[[(1R,3R,4R,7S)-7-[[(2R,3S,5R)-5-(6-aminopurin-9-yl)-3-[[(2R,3S,5R)-5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3-oxidanyl-oxolan-2-yl]methoxy-sulfanyl-phosphoryl]oxy-oxolan-2-yl]methoxy-sulfanyl-phosphoryl]oxy-3-(4-azanyl-5-methyl-2-oxidanylidene-pyrimidin-1-yl)-2,5-dioxabicyclo[2.2.1]heptan-1-yl]methoxy-sulfanyl-phosphoryl]oxy-5-(hydroxymethyl)oxolan-2-yl]-5-methyl-pyrimidine-2,4-dione, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Hung-En, H, Zanini, C, Simonneau, C, Fraidling, J, Kraft, T, Mayer, K, Sommer, A, Indlekofer, A, Wirth, T, Benz, J, Georges, G, Langer, M, Gassner, C, Larraillet, V, Manso, M, Ravn, J, Hofer, K, Emrich, T, Niewoehner, J, Schumacher, F, Brinkmann, U. | | Deposit date: | 2024-08-08 | | Release date: | 2025-08-20 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Improved Targeted Delivery of Antisense Oligonucleotide Conjugates
with the Antibody Mask
To Be Published
|
|
9GFP
 
 | | The crystal structure of CsvR from Escherichia coli | | Descriptor: | HTH-type transcriptional activator CsvR | | Authors: | Moche, M, Joffre, E, Ampah-Korsah, H, Nyman, T, Sjoling, A, Sun, L. | | Deposit date: | 2024-08-12 | | Release date: | 2025-08-20 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | The crystal structure of CsvR from Escherichia coli
To Be Published
|
|
2PCD
 
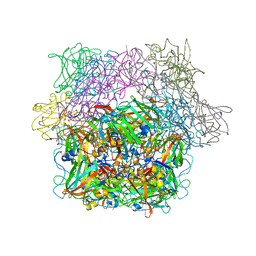 | | STRUCTURE OF PROTOCATECHUATE 3,4-DIOXYGENASE FROM PSEUDOMONAS AERUGINOSA AT 2.15 ANGSTROMS RESOLUTION | | Descriptor: | FE (III) ION, PROTOCATECHUATE 3,4-DIOXYGENASE (ALPHA CHAIN), PROTOCATECHUATE 3,4-DIOXYGENASE (BETA CHAIN) | | Authors: | Ohlendorf, D.H, Orville, A.M, Lipscomb, J.D. | | Deposit date: | 1994-06-21 | | Release date: | 1994-12-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of protocatechuate 3,4-dioxygenase from Pseudomonas aeruginosa at 2.15 A resolution.
J.Mol.Biol., 244, 1994
|
|
1MHZ
 
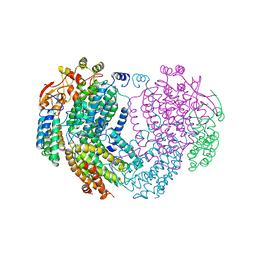 | | METHANE MONOOXYGENASE HYDROXYLASE | | Descriptor: | FE (III) ION, METHANE MONOOXYGENASE HYDROXYLASE | | Authors: | Elango, N, Radhakrishnan, R, Froland, W.A, Waller, B.J, Earhart, C.A, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 1996-10-21 | | Release date: | 1997-05-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the hydroxylase component of methane monooxygenase from Methylosinus trichosporium OB3b
Protein Sci., 6, 1997
|
|
9GGA
 
 | |
1MP5
 
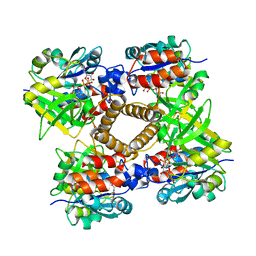 | | Y177F VARIANT OF S. ENTERICA RmlA | | Descriptor: | URIDINE-5'-DIPHOSPHATE-GLUCOSE, Y177F VARIANT OF S. ENTERICA RmlA BOUND TO UDP-GLUCOSE | | Authors: | Barton, W.A, Biggins, J.B, Jiang, J, Thorson, J.S, Nikolov, D.B. | | Deposit date: | 2002-09-11 | | Release date: | 2002-10-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Expanding pyrimidine diphosphosugar libraries via structure-based nucleotidylyltransferase engineering
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
9GI1
 
 | | Structure of the S.aureus MecA/ClpC/ClpP degradation system | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpC, ATP-dependent Clp protease proteolytic subunit, ... | | Authors: | Azinas, S, Wallden, K, Katikaridis, P, Schahl, A, Mogk, A, Carroni, M. | | Deposit date: | 2024-08-16 | | Release date: | 2025-08-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | ClpC and ClpP act as reciprocal allosteric activators to form a highly efficient AAA+ protease
To Be Published
|
|
1MQ4
 
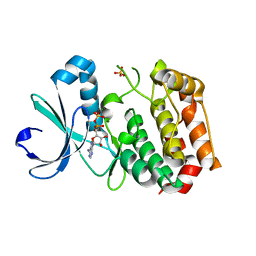 | | Crystal Structure of Aurora-A Protein Kinase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, AURORA-RELATED KINASE 1, MAGNESIUM ION, ... | | Authors: | Nowakowski, J, Cronin, C.N, McRee, D.E, Knuth, M.W, Nelson, C, Pavletich, N.P, Rodgers, J, Sang, B.-C, Scheibe, D.N, Swanson, R.V, Thompson, D.A. | | Deposit date: | 2002-09-13 | | Release date: | 2003-09-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of the Cancer-Related Aurora-A, FAK and EphA2 Protein Kinases from Nanovolume Crystallography
Structure, 10, 2002
|
|
9GHK
 
 | |
9GS0
 
 | | Capsid of full Haloferax tailed virus 1 without turret head protein gp31. | | Descriptor: | Capsid stabilization protein, Gp30, HK97 gp5-like major capsid protein, ... | | Authors: | Zhang, D, Daum, B, Isupov, M.N, McLaren, M, Stuart, W. | | Deposit date: | 2024-09-13 | | Release date: | 2025-08-27 | | Last modified: | 2025-10-22 | | Method: | ELECTRON MICROSCOPY (2.37 Å) | | Cite: | Cryo-EM resolves the structure of the archaeal dsDNA virus HFTV1 from head to tail.
Sci Adv, 11, 2025
|
|
1MIZ
 
 | | Crystal structure of an integrin beta3-talin chimera | | Descriptor: | TALIN, integrin beta3 | | Authors: | Garcia-Alvarez, B, de Pereda, J.M, Calderwood, D.A, Ulmer, T.S, Critchley, D, Campbell, I.D, Ginsberg, M.H, Liddington, R.C. | | Deposit date: | 2002-08-23 | | Release date: | 2003-01-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural determinants of integrin recognition by talin
Mol.Cell, 11, 2003
|
|
9GFJ
 
 | | Crystal structure of ASO binding Fab fragment with ASO143 | | Descriptor: | 1,2-ETHANEDIOL, 1-[(1~{S},3~{R},4~{R},7~{S})-7-[[(1~{R},3~{R},4~{R},7~{S})-7-[[(1~{R},3~{R},4~{R},7~{S})-3-(6-aminopurin-9-yl)-7-[[(1~{R},3~{R},4~{R},7~{S})-3-(4-azanyl-5-methyl-2-oxidanylidene-pyrimidin-1-yl)-7-oxidanyl-2,5-dioxabicyclo[2.2.1]heptan-1-yl]methoxy-sulfanyl-phosphoryl]oxy-2,5-dioxabicyclo[2.2.1]heptan-1-yl]methoxy-sulfanyl-phosphoryl]oxy-3-(4-azanyl-5-methyl-2-oxidanylidene-pyrimidin-1-yl)-2,5-dioxabicyclo[2.2.1]heptan-1-yl]methoxy-sulfanyl-phosphoryl]oxy-1-(hydroxymethyl)-2,5-dioxabicyclo[2.2.1]heptan-3-yl]-5-methyl-pyrimidine-2,4-dione, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Hsia, H.-E, Zanini, C, Simonneau, C, Fraidling, J, Kraft, T, Mayer, K, Sommer, A, Indlekofer, A, Wirht, T, Benz, J, Georges, G, Langer, L.M, Gassner, C, Larraillet, V, Manso, M, Ravn, J, Hofer, K, Emrich, T, Niewoehner, J, Schumacher, F, Brinkmann, U. | | Deposit date: | 2024-08-09 | | Release date: | 2025-08-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Improved Targeted Delivery of Antisense Oligonucleotide Conjugates
with the Antibody Mask
To Be Published
|
|
1MJ8
 
 | | High Resolution Crystal Structure Of The Fab Fragment of The Esterolytic Antibody MS6-126 | | Descriptor: | GLYCEROL, IMMUNOGLOBULIN MS6-126, PHOSPHATE ION | | Authors: | Ruzheinikov, S.N, Muranova, T.A, Sedelnikova, S.E, Partridge, L.J, Blackburn, G.M, Murray, I.A, Kakinuma, H, Takashi, N, Shimazaki, K, Sun, J, Nishi, Y, Rice, D.W. | | Deposit date: | 2002-08-27 | | Release date: | 2003-09-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | High-resolution crystal structure of the Fab-fragments of a family of mouse catalytic antibodies with esterase activity
J.Mol.Biol., 332, 2003
|
|
