3L6E
 
 | | Crystal structure of putative short chain dehydrogenase/reductase family oxidoreductase from Aeromonas hydrophila subsp. hydrophila ATCC 7966 | | Descriptor: | Oxidoreductase, short-chain dehydrogenase/reductase family, SULFATE ION | | Authors: | Malashkevich, V.N, Patskovsky, Y, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-12-23 | | Release date: | 2010-02-09 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of putative short chain dehydrogenase/reductase family oxidoreductase from Aeromonas
hydrophila subsp. hydrophila ATCC 7966
To be Published
|
|
3L6T
 
 | |
3LMU
 
 | | Crystal structure of DTD from Plasmodium falciparum | | Descriptor: | D-tyrosyl-tRNA(Tyr) deacylase, IODIDE ION | | Authors: | Manickam, Y, Bhatt, T.K, Khan, S, Sharma, A. | | Deposit date: | 2010-02-01 | | Release date: | 2010-03-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of D-tyrosyl-tRNATyr deacylase using home-source Cu Kalpha and moderate-quality iodide-SAD data: structural polymorphism and HEPES-bound enzyme states
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3L9B
 
 | | Crystal Structure of Rat Otoferlin C2A | | Descriptor: | MAGNESIUM ION, Otoferlin | | Authors: | Helfmann, S, Neumann, P. | | Deposit date: | 2010-01-04 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The crystal structure of the C2A domain of otoferlin reveals an unconventional top loop region.
J.Mol.Biol., 406, 2011
|
|
3L9Z
 
 | | Crystal Structure of UreE from Helicobacter pylori (apo form) | | Descriptor: | Urease accessory protein ureE | | Authors: | Shi, R, Munger, C, Assinas, A, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2010-01-06 | | Release date: | 2010-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal Structures of Apo and Metal-Bound Forms of the UreE Protein from Helicobacter pylori: Role of Multiple Metal Binding Sites
Biochemistry, 49, 2010
|
|
3LAA
 
 | |
3LB5
 
 | |
3L5U
 
 | |
2Q8N
 
 | |
3P1U
 
 | |
3OZF
 
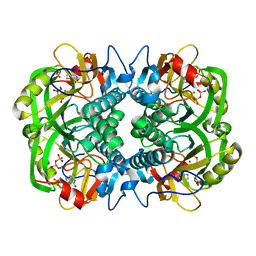 | | Crystal Structure of Plasmodium falciparum Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferase in complex with hypoxanthine | | Descriptor: | HYPOXANTHINE, Hypoxanthine-guanine-xanthine phosphoribosyltransferase, MAGNESIUM ION, ... | | Authors: | Ho, M, Hazleton, K.Z, Almo, S.C, Schramm, V.L. | | Deposit date: | 2010-09-24 | | Release date: | 2011-09-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.944 Å) | | Cite: | Acyclic Immucillin Phosphonates: Second-Generation Inhibitors of Plasmodium falciparum Hypoxanthine- Guanine-Xanthine Phosphoribosyltransferase.
Chem.Biol., 19, 2012
|
|
3P0J
 
 | |
3P1E
 
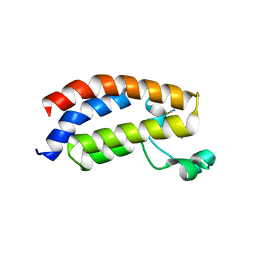 | | Crystal structure of the bromodomain of human CREBBP in complex with dimethyl sulfoxide (DMSO) | | Descriptor: | CREB-binding protein, DIMETHYL SULFOXIDE, POTASSIUM ION, ... | | Authors: | Filippakopoulos, P, Picaud, S, Feletar, I, Fedorov, O, Muniz, J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-09-30 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the bromodomain of human CREBBP in complex with dimethyl sulfoxide (DMSO)
To be Published
|
|
3P54
 
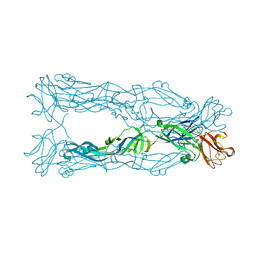 | | Crystal Structure of the Japanese Encephalitis Virus Envelope Protein, strain SA-14-14-2. | | Descriptor: | envelope glycoprotein | | Authors: | Luca, V.C, Nelson, C.A, AbiMansour, J.P, Diamond, M.S, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-10-07 | | Release date: | 2010-12-08 | | Last modified: | 2012-02-08 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Crystal structure of the Japanese encephalitis virus envelope protein.
J.Virol., 86, 2012
|
|
1ZKZ
 
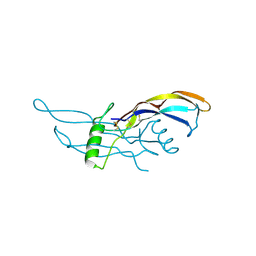 | | Crystal Structure of BMP9 | | Descriptor: | Growth/differentiation factor 2 | | Authors: | Brown, M.A, Zhao, Q, Baker, K.A, Naik, C, Chen, C, Pukac, L, Singh, M, Tsareva, T, Parice, Y, Mahoney, A, Roschke, V, Sanyal, I, Choe, S. | | Deposit date: | 2005-05-04 | | Release date: | 2005-05-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure of BMP-9 and functional interactions with pro-region and receptors
J.Biol.Chem., 280, 2005
|
|
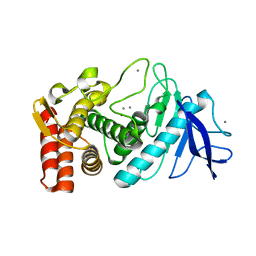
3P7V
 
 | |
3P9L
 
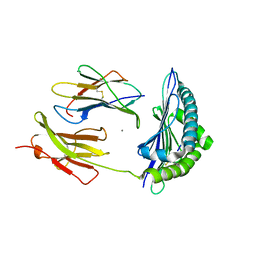 | | Crystal Structure of H2-Kb in complex with the chicken ovalbumin epitope OVA | | Descriptor: | Beta-2-microglobulin, CALCIUM ION, H-2 class I histocompatibility antigen, ... | | Authors: | Wesselingh, R, Gras, S, Guillonneau, C, Turner, S.J, Rossjohn, J. | | Deposit date: | 2010-10-18 | | Release date: | 2011-10-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Affinity thresholds for naive CD8+ CTL activation by peptides and engineered influenza A viruses
J.Immunol., 187, 2011
|
|
3P0E
 
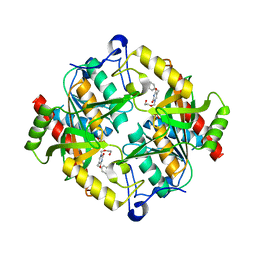 | | Structure of hUPP2 in an active conformation with bound 5-benzylacyclouridine | | Descriptor: | 1-((2-HYDROXYETHOXY)METHYL)-5-BENZYLPYRIMIDINE-2,4(1H,3H)-DIONE, PHOSPHATE ION, Uridine phosphorylase 2 | | Authors: | Roosild, T.P, Castronovo, S, Villoso, A. | | Deposit date: | 2010-09-28 | | Release date: | 2011-09-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A novel structural mechanism for redox regulation of uridine phosphorylase 2 activity.
J.Struct.Biol., 176, 2011
|
|
3PAY
 
 | |
3P1T
 
 | |
3P5M
 
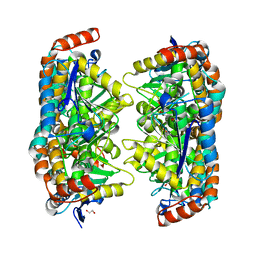 | |
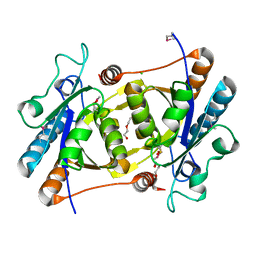
3OQH
 
 | | Crystal structure of B. licheniformis CDPS yvmC-BLIC | | Descriptor: | GLYCEROL, Putative uncharacterized protein yvmC | | Authors: | Bonnefond, L, Arai, T, Suzuki, T, Ishitani, R, Nureki, O. | | Deposit date: | 2010-09-03 | | Release date: | 2011-02-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structural basis for nonribosomal peptide synthesis by an aminoacyl-tRNA synthetase paralog.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3OQP
 
 | |
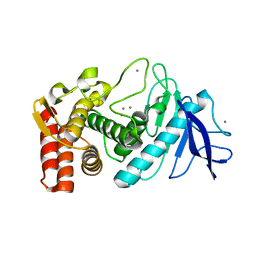
3P7T
 
 | |
3ORE
 
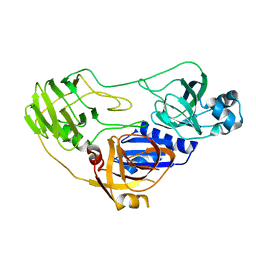 | | Crystal structure of TTHA0988 in space group P6522 | | Descriptor: | Putative uncharacterized protein TTHA0988 | | Authors: | Jacques, D.A, Kuramitsu, S, Yokoyama, S, Trewhella, J, Guss, J.M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-09-07 | | Release date: | 2011-02-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of TTHA0988 from Thermus thermophilus, a KipI-KipA homologue incorrectly annotated as an allophanate hydrolase
Acta Crystallogr.,Sect.D, 67, 2011
|
|
