1KZG
 
 | | DbsCdc42(Y889F) | | Descriptor: | CDC42 HOMOLOG, GUANINE NUCLEOTIDE EXCHANGE FACTOR DBS | | Authors: | Rossman, K.L, Worthylake, D.K, Snyder, J.T, Siderovski, D.P, Campbell, S.L, Sondek, J. | | Deposit date: | 2002-02-06 | | Release date: | 2002-03-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A crystallographic view of interactions between Dbs and Cdc42: PH domain-assisted guanine nucleotide exchange.
EMBO J., 21, 2002
|
|
7PQH
 
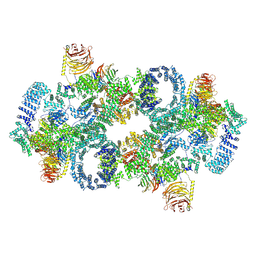 | | Cryo-EM structure of Saccharomyces cerevisiae TOROID (TORC1 Organized in Inhibited Domains). | | Descriptor: | Serine/threonine-protein kinase TOR2, Target of rapamycin complex 1 subunit KOG1,Target of rapamycin complex 1 subunit Kog1, Target of rapamycin complex subunit LST8 | | Authors: | Felix, J, Prouteau, M, Bourgoint, C, Bonadei, L, Desfosses, A, Gabus, C, Sadian, Y, Savvides, S.N, Gutsche, I, Loewith, R. | | Deposit date: | 2021-09-17 | | Release date: | 2023-01-18 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | EGOC inhibits TOROID polymerization by structurally activating TORC1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
1MYO
 
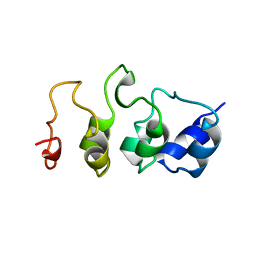 | | SOLUTION STRUCTURE OF MYOTROPHIN, NMR, 44 STRUCTURES | | Descriptor: | MYOTROPHIN | | Authors: | Yang, Y, Nanduri, S, Sen, S, Qin, J. | | Deposit date: | 1998-08-17 | | Release date: | 1999-08-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structural basis of ankyrin-like repeat function as revealed by the solution structure of myotrophin.
Structure, 6, 1998
|
|
2BJR
 
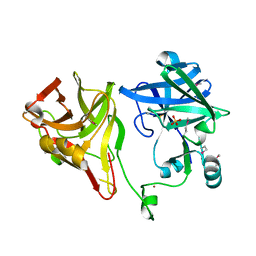 | | Crystal structure of the nematode sperm cell motility protein MFP2B | | Descriptor: | MFP2B, SULFATE ION, ZINC ION | | Authors: | Grant, R.P, Buttery, S.M, Ekman, G.C, Roberts, T.M, Stewart, M. | | Deposit date: | 2005-02-07 | | Release date: | 2005-03-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Mfp2 and its Function in Enhancing Msp Polymerization in Ascaris Sperm Amoeboid Motility
J.Mol.Biol., 347, 2005
|
|
3QWX
 
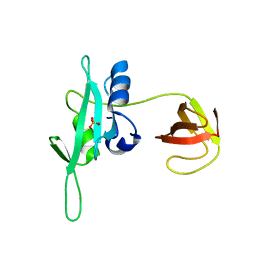 | | CED-2 1-174 | | Descriptor: | Cell death abnormality protein 2, SULFATE ION | | Authors: | Kang, Y, Sun, J, Liu, Y. | | Deposit date: | 2011-02-28 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of the cell corpse engulfment protein CED-2 in Caenorhabditis elegans.
Biochem.Biophys.Res.Commun., 410, 2011
|
|
1LVK
 
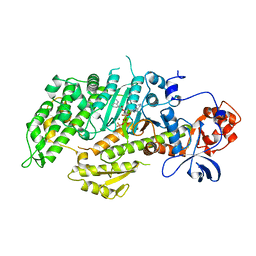 | | X-RAY CRYSTAL STRUCTURE OF THE MG (DOT) 2'(3')-O-(N-METHYLANTHRANILOYL) NUCLEOTIDE BOUND TO DICTYOSTELIUM DISCOIDEUM MYOSIN MOTOR DOMAIN | | Descriptor: | 2'(3')-O-N-METHYLANTHRANILOYL-ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Bauer, C.B, Kuhlman, P.A, Bagshaw, C.R, Rayment, I. | | Deposit date: | 1997-09-05 | | Release date: | 1998-01-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray crystal structure and solution fluorescence characterization of Mg.2'(3')-O-(N-methylanthraniloyl) nucleotides bound to the Dictyostelium discoideum myosin motor domain.
J.Mol.Biol., 274, 1997
|
|
1WYP
 
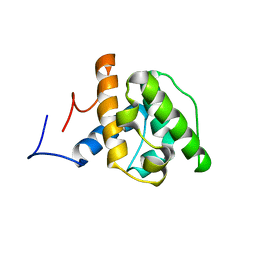 | | Solution structure of the CH domain of human Calponin 1 | | Descriptor: | Calponin 1 | | Authors: | Tomizawa, T, Kigawa, T, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-02-15 | | Release date: | 2005-08-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the CH domain of human Calponin 1
To be Published
|
|
1KZ7
 
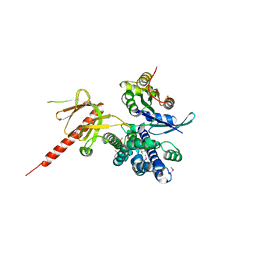 | | Crystal Structure of the DH/PH Fragment of Murine Dbs in Complex with the Placental Isoform of Human Cdc42 | | Descriptor: | CDC42 HOMOLOG, GUANINE NUCLEOTIDE EXCHANGE FACTOR DBS | | Authors: | Rossman, K.L, Worthylake, D.K, Snyder, J.T, Siderovski, D.P, Campbell, S.L, Sondek, J. | | Deposit date: | 2002-02-06 | | Release date: | 2002-03-20 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A crystallographic view of interactions between Dbs and Cdc42: PH domain-assisted guanine nucleotide exchange.
EMBO J., 21, 2002
|
|
7SCS
 
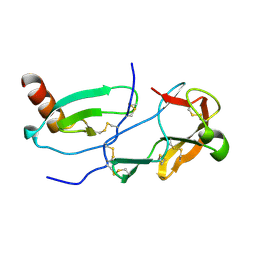 | | Crystal Structure of the Tick Evasin EVA-AAM1001 Complexed to Human Chemokine CCL11 | | Descriptor: | Eotaxin, Evasin P1243 | | Authors: | Devkota, S.R, Bhusal, R.P, Aryal, P, Wilce, M.C.J, Stone, M.J. | | Deposit date: | 2021-09-29 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Engineering broad-spectrum inhibitors of inflammatory chemokines from subclass A3 tick evasins.
Nat Commun, 14, 2023
|
|
1MND
 
 | |
3S28
 
 | | The crystal structure of sucrose synthase-1 in complex with a breakdown product of the UDP-glucose | | Descriptor: | 1,5-anhydro-D-arabino-hex-1-enitol, 1,5-anhydro-D-fructose, MALONIC ACID, ... | | Authors: | Zheng, Y, Garavito, R.M. | | Deposit date: | 2011-05-16 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Structure of Sucrose Synthase-1 from Arabidopsis thaliana and Its Functional Implications.
J.Biol.Chem., 286, 2011
|
|
1B4S
 
 | | STRUCTURE OF NUCLEOSIDE DIPHOSPHATE KINASE H122G MUTANT | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, NUCLEOSIDE DIPHOSPHATE KINASE, ... | | Authors: | Meyer, P, Janin, J. | | Deposit date: | 1998-12-28 | | Release date: | 1999-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Nucleophilic activation by positioning in phosphoryl transfer catalyzed by nucleoside diphosphate kinase.
Biochemistry, 38, 1999
|
|
6V69
 
 | | Structures of GCP4 and GCP5 in the native human gamma-tubulin ring complex | | Descriptor: | Gamma-tubulin complex component 4, Gamma-tubulin complex component 5 | | Authors: | Wieczorek, M, Urnavicius, L, Ti, S, Molloy, K.R, Chait, B.T, Kapoor, T.M. | | Deposit date: | 2019-12-04 | | Release date: | 2020-01-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Asymmetric Molecular Architecture of the Human gamma-Tubulin Ring Complex.
Cell, 180, 2020
|
|
6V6C
 
 | | Structure of GCP6 in the native human gamma-tubulin ring complex | | Descriptor: | Gamma-tubulin complex component 6 | | Authors: | Wieczorek, M, Urnavicius, L, Ti, S, Molloy, K.R, Chait, B.T, Kapoor, T.M. | | Deposit date: | 2019-12-04 | | Release date: | 2020-01-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Asymmetric Molecular Architecture of the Human gamma-Tubulin Ring Complex.
Cell, 180, 2020
|
|
6V5V
 
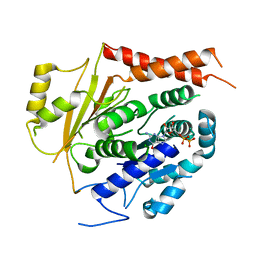 | | Structure of gamma-tubulin in the native human gamma-tubulin ring complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Tubulin gamma-1 chain | | Authors: | Wieczorek, M, Urnavicius, L, Ti, S, Molloy, K.R, Chait, B.T, Kapoor, T.M. | | Deposit date: | 2019-12-04 | | Release date: | 2020-01-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Asymmetric Molecular Architecture of the Human gamma-Tubulin Ring Complex.
Cell, 180, 2020
|
|
6V6B
 
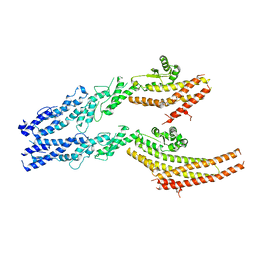 | | Structures of GCP2 and GCP3 in the native human gamma-tubulin ring complex | | Descriptor: | Gamma-tubulin complex component 2, Gamma-tubulin complex component 3 | | Authors: | Wieczorek, M, Urnavicius, L, Ti, S, Molloy, K.R, Chait, B.T, Kapoor, T.M. | | Deposit date: | 2019-12-04 | | Release date: | 2020-01-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Asymmetric Molecular Architecture of the Human gamma-Tubulin Ring Complex.
Cell, 180, 2020
|
|
1E0S
 
 | | small G protein Arf6-GDP | | Descriptor: | ADP-ribosylation factor 6, AMMONIUM ION, BETA-MERCAPTOETHANOL, ... | | Authors: | Menetrey, J, Cherfils, J. | | Deposit date: | 2000-04-06 | | Release date: | 2000-04-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structure of Arf6-Gdp Suggests a Basis for Guanine Nucleotide Exchange Factors Specificity
Nat.Struct.Biol., 7, 2000
|
|
1EOT
 
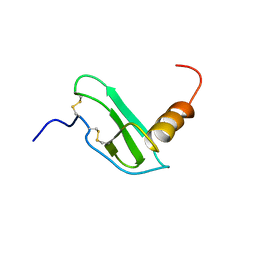 | |
1BUX
 
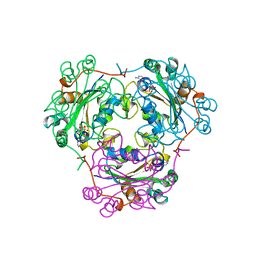 | | 3'-PHOSPHORYLATED NUCLEOTIDES BINDING TO NUCLEOSIDE DIPHOSPHATE KINASE | | Descriptor: | 3'-PHOSPHATE-ADENOSINE-5'-PHOSPHATE SULFATE, NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Xu, Y, Schneider, B, Deville-Bonne, D, Veron, M, Janin, J. | | Deposit date: | 1998-09-07 | | Release date: | 1999-04-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 3'-Phosphorylated nucleotides are tight binding inhibitors of nucleoside diphosphate kinase activity.
J.Biol.Chem., 273, 1998
|
|
1DG3
 
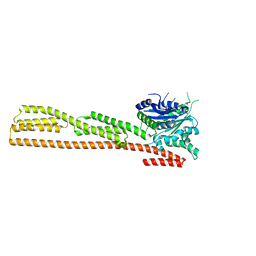 | | STRUCTURE OF HUMAN GUANYLATE BINDING PROTEIN-1 IN NUCLEOTIDE FREE FORM | | Descriptor: | PROTEIN (INTERFERON-INDUCED GUANYLATE-BINDING PROTEIN 1) | | Authors: | Prakash, B, Praefcke, G.J.K, Renault, L, Wittinghofer, A, Herrmann, C. | | Deposit date: | 1999-11-23 | | Release date: | 2000-10-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of human guanylate-binding protein 1 representing a unique class of GTP-binding proteins.
Nature, 403, 2000
|
|
1CD9
 
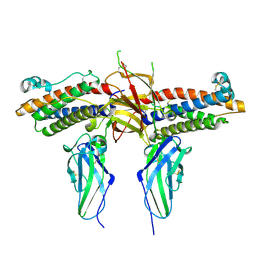 | | 2:2 COMPLEX OF G-CSF WITH ITS RECEPTOR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (G-CSF RECEPTOR), PROTEIN (GRANULOCYTE COLONY-STIMULATING FACTOR) | | Authors: | Aritomi, M, Kunishima, N, Okamoto, T, Kuroki, R, Ota, Y, Morikawa, K. | | Deposit date: | 1999-03-08 | | Release date: | 2000-03-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Atomic structure of the GCSF-receptor complex showing a new cytokine-receptor recognition scheme.
Nature, 401, 1999
|
|
1F3F
 
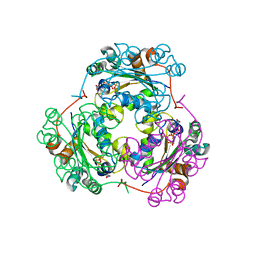 | | STRUCTURE OF THE H122G NUCLEOSIDE DIPHOSPHATE KINASE / D4T-TRIPHOSPHATE.MG COMPLEX | | Descriptor: | 2',3'-DEHYDRO-2',3'-DEOXY-THYMIDINE 5'-DIPHOSPHATE, 2',3'-DEHYDRO-2',3'-DEOXY-THYMIDINE 5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Meyer, P, Schneider, B, Sarfati, S, Deville-Bonne, D, Guerreiro, C, Boretto, J, Janin, J, Veron, M, Canard, B. | | Deposit date: | 2000-06-02 | | Release date: | 2000-09-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for activation of alpha-boranophosphate nucleotide analogues targeting drug-resistant reverse transcriptase.
EMBO J., 19, 2000
|
|
1F5N
 
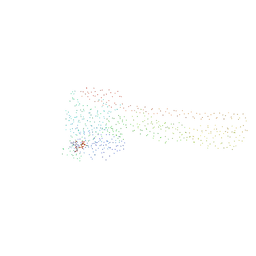 | | HUMAN GUANYLATE BINDING PROTEIN-1 IN COMPLEX WITH THE GTP ANALOGUE, GMPPNP. | | Descriptor: | INTERFERON-INDUCED GUANYLATE-BINDING PROTEIN 1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Prakash, B, Renault, L, Praefcke, G.J.K, Herrmann, C, Wittinghofer, A. | | Deposit date: | 2000-06-15 | | Release date: | 2000-09-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Triphosphate structure of guanylate-binding protein 1 and implications for nucleotide binding and GTPase mechanism.
EMBO J., 19, 2000
|
|
1F6T
 
 | | STRUCTURE OF THE NUCLEOSIDE DIPHOSPHATE KINASE/ALPHA-BORANO(RP)-TDP.MG COMPLEX | | Descriptor: | 2*-DEOXY-THYMIDINE-5*-ALPHA BORANO DIPHOSPHATE (ISOMER RP), MAGNESIUM ION, PROTEIN (NUCLEOSIDE DIPHOSPHATE KINASE) | | Authors: | Guerreiro, C, Boretto, J, Janin, J, Veron, M, Canard, B, Schneider, B, Sarfati, S, Deville-Bonne, D. | | Deposit date: | 2000-06-23 | | Release date: | 2000-09-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural basis for activation of alpha-boranophosphate nucleotide analogues targeting drug-resistant reverse transcriptase.
EMBO J., 19, 2000
|
|
1B99
 
 | | 3'-FLUORO-URIDINE DIPHOSPHATE BINDING TO NUCLEOSIDE DIPHOSPHATE KINASE | | Descriptor: | 2',3'-DIDEOXY-3'-FLUORO-URIDIDINE-5'-DIPHOSPHATE, PROTEIN (NUCLEOSIDE DIPHOSPHATE KINASE), PYROPHOSPHATE 2- | | Authors: | Janin, J, Xu, Y. | | Deposit date: | 1999-02-22 | | Release date: | 1999-06-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Catalytic mechanism of nucleoside diphosphate kinase investigated using nucleotide analogues, viscosity effects, and X-ray crystallography.
Biochemistry, 38, 1999
|
|
