2VTI
 
 | | Identification of N-(4-piperidinyl)-4-(2,6-dichlorobenzoylamino)-1H- pyrazole-3-carboxamide (AT7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design. | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, N-(4-sulfamoylphenyl)-1H-indazole-3-carboxamide | | Authors: | Wyatt, P.G, Woodhead, A.J, Boulstridge, J.A, Berdini, V, Carr, M.G, Cross, D.M, Danillon, D, Davis, D.J, Devine, L.A, Early, T.R, Feltell, R.E, Lewis, E.J, McMenamin, R.L, Navarro, E.F, O'Brien, M.A, O'Reilly, M, Reule, M, Saxty, G, Seavers, L.C.A, Smith, D, Squires, M.S, Trewartha, G, Walker, M.T, Woolford, A.J. | | Deposit date: | 2008-05-15 | | Release date: | 2008-08-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of N-(4-Piperidinyl)-4-(2,6-Dichlorobenzoylamino)-1H-Pyrazole-3-Carboxamide (at7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design.
J.Med.Chem., 51, 2008
|
|
2WHB
 
 | | Truncation and Optimisation of Peptide Inhibitors of CDK2, Cyclin A Through Structure Guided Design | | Descriptor: | ARG-ARG-L3O-PFF, CELL DIVISION PROTEIN KINASE 2, CYCLIN-A2 | | Authors: | Kontopidis, G, Andrews, M.J, McInnes, C, Plater, A, Innes, L, Renachowski, S, Cowan, A, Fischer, P.M. | | Deposit date: | 2009-05-03 | | Release date: | 2009-06-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Truncation and optimisation of peptide inhibitors of cyclin-dependent kinase 2-cyclin a through structure-guided design.
Chemmedchem, 4, 2009
|
|
5FCW
 
 | | HDAC8 Complexed with a Hydroxamic Acid | | Descriptor: | 4-naphthalen-1-yl-~{N}-oxidanyl-benzamide, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Cole, K.E, Perry, K. | | Deposit date: | 2015-12-15 | | Release date: | 2016-10-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.979 Å) | | Cite: | Structure of 'linkerless' hydroxamic acid inhibitor-HDAC8 complex confirms the formation of an isoform-specific subpocket.
J.Struct.Biol., 195, 2016
|
|
2WMA
 
 | |
5G1C
 
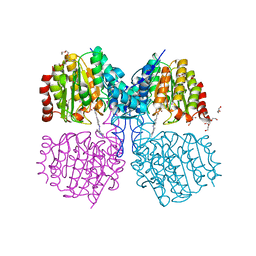 | | Structure of HDAC like protein from Bordetella Alcaligenes bound the photoswitchable pyrazole Inhibitor CEW395 | | Descriptor: | (2E)-N-hydroxy-3-{4-[(E)-(1,3,5-trimethyl-1H-pyrazol-4-yl)diazenyl]phenyl}prop-2-enamide, DI(HYDROXYETHYL)ETHER, HISTONE DEACETYLASE-LIKE AMIDOHYDROLASE, ... | | Authors: | Kraemer, A, Meyer-Almes, F.J, Yildiz, O. | | Deposit date: | 2016-03-24 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Toward Photopharmacological Antimicrobial Chemotherapy Using Photoswitchable Amidohydrolase Inhibitors.
ACS Infect Dis, 3, 2017
|
|
6HSK
 
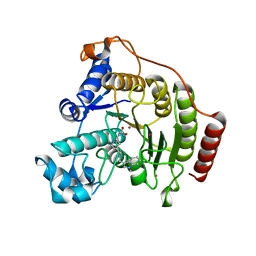 | | Crystal structure of a human HDAC8 L6 loop mutant complexed with Quisinostat | | Descriptor: | 2-[4-[[(1-methylindol-3-yl)methylamino]methyl]piperidin-1-yl]-~{N}-oxidanyl-pyrimidine-5-carboxamide, Histone deacetylase 8, POTASSIUM ION, ... | | Authors: | Marek, M, Shaik, T.B, Ramos-Morales, E, Romier, C. | | Deposit date: | 2018-10-01 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
6HSZ
 
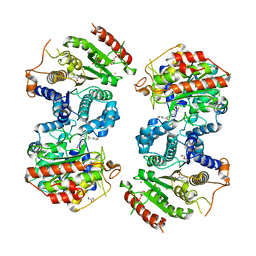 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with a benzohydroxamate inhibitor 2 | | Descriptor: | 3-benzamido-4-methyl-~{N}-oxidanyl-benzamide, GLYCEROL, Histone deacetylase, ... | | Authors: | Marek, M, Shaik, T.B, Romier, C. | | Deposit date: | 2018-10-02 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.374 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
6HRQ
 
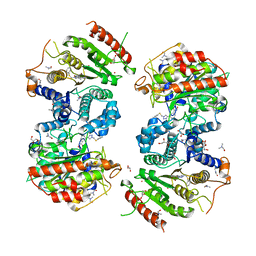 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with NCC-149 | | Descriptor: | DIMETHYLFORMAMIDE, GLYCEROL, Histone deacetylase, ... | | Authors: | Shaik, T.B, Marek, M, Romier, C. | | Deposit date: | 2018-09-28 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.845 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
6HTI
 
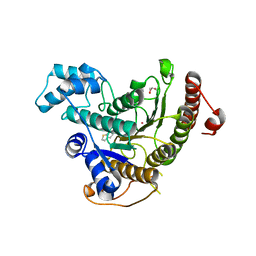 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with a benzohydroxamate inhibitor 6 | | Descriptor: | GLYCEROL, Histone deacetylase, POTASSIUM ION, ... | | Authors: | Shaik, T.B, Marek, M, Romier, C. | | Deposit date: | 2018-10-04 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.693 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
6HTT
 
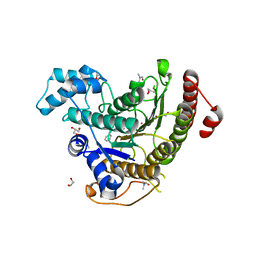 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with a benzohydroxamate inhibitor 7 | | Descriptor: | DIMETHYLFORMAMIDE, GLYCEROL, Histone deacetylase, ... | | Authors: | Shaik, T.B, Marek, M, Romier, C. | | Deposit date: | 2018-10-04 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.748 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
6XTB
 
 | |
6E3K
 
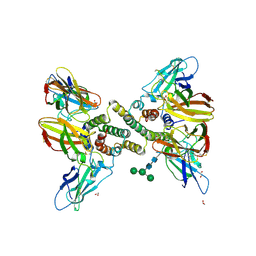 | | Interferon gamma signalling complex with IFNGR1 and IFNGR2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jude, K.M, Mendoza, J.L, Garcia, K.C. | | Deposit date: | 2018-07-14 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure of the IFN gamma receptor complex guides design of biased agonists.
Nature, 567, 2019
|
|
6HSF
 
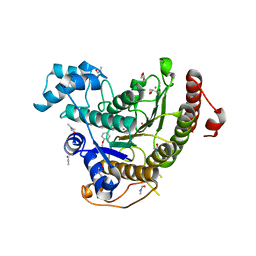 | | Crystal structure of Schistosoma mansoni HDAC8 mutant H292M complexed with PCI-34051 | | Descriptor: | 1-[(4-methoxyphenyl)methyl]-~{N}-oxidanyl-indole-6-carboxamide, DIMETHYLFORMAMIDE, GLYCEROL, ... | | Authors: | Shaik, T.B, Marek, M, Romier, C. | | Deposit date: | 2018-10-01 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
6HTH
 
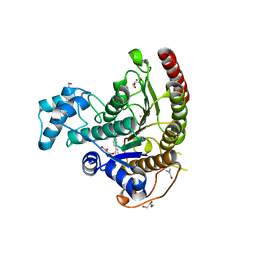 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with a benzohydroxamate inhibitor 5 | | Descriptor: | 4-methoxy-~{N}-oxidanyl-3-[(4-phenylphenyl)carbonylamino]benzamide, DIMETHYLFORMAMIDE, GLYCEROL, ... | | Authors: | Shaik, T.B, Marek, M, Romier, C. | | Deposit date: | 2018-10-04 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
6HU3
 
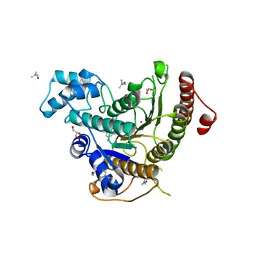 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with a triazole hydroxamate inhibitor | | Descriptor: | 1-[5-chloranyl-2-(4-fluoranylphenoxy)phenyl]-~{N}-oxidanyl-1,2,3-triazole-4-carboxamide, DIMETHYLFORMAMIDE, GLYCEROL, ... | | Authors: | Shaik, T.B, Marek, M, Romier, C. | | Deposit date: | 2018-10-05 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.655 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
6HSH
 
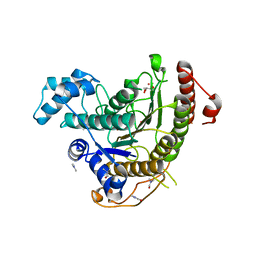 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with Quisinostat | | Descriptor: | 2-[4-[[(1-methylindol-3-yl)methylamino]methyl]piperidin-1-yl]-~{N}-oxidanyl-pyrimidine-5-carboxamide, DIMETHYLFORMAMIDE, GLYCEROL, ... | | Authors: | Shaik, T.B, Marek, M, Romier, C. | | Deposit date: | 2018-10-01 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.545 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
6HT8
 
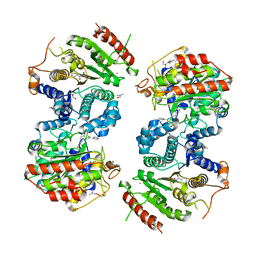 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with a benzohydroxamate inhibitor 3 | | Descriptor: | 3-benzamido-4-methoxy-~{N}-oxidanyl-benzamide, DIMETHYLFORMAMIDE, GLYCEROL, ... | | Authors: | Marek, M, Shaik, T.B, Romier, C. | | Deposit date: | 2018-10-03 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
6HT5
 
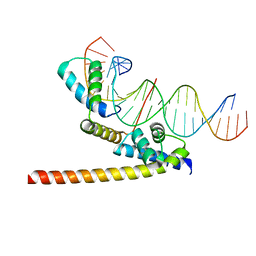 | | Oct4/Sox2:UTF1 structure | | Descriptor: | DNA (5'-D(*TP*TP*CP*AP*CP*TP*AP*GP*CP*AP*TP*AP*AP*CP*AP*AP*TP*GP*AP*GP*G)-3'), DNA (5'-D(P*CP*CP*TP*CP*AP*TP*TP*GP*TP*TP*AP*TP*GP*CP*TP*AP*GP*TP*GP*AP*AP*G)-3'), POU domain, ... | | Authors: | Vahokoski, J, Meusch, D, Groves, M, Pogenberg, V, Wilmanns, M. | | Deposit date: | 2018-10-03 | | Release date: | 2019-10-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.451 Å) | | Cite: | Oct4/Sox2:UTF1 structure
To Be Published
|
|
6HU2
 
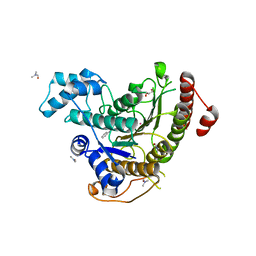 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with a benzohydroxamate inhibitor 11 | | Descriptor: | 4-methyl-~{N}-oxidanyl-3-[(phenylmethyl)amino]benzamide, DIMETHYLFORMAMIDE, GLYCEROL, ... | | Authors: | Shaik, T.B, Marek, M, Romier, C. | | Deposit date: | 2018-10-05 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.986 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
5G49
 
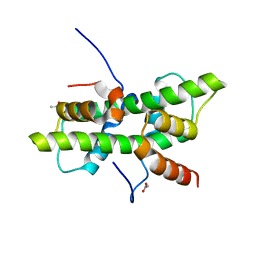 | | Crystal structure of the Arabodopsis thaliana histone-fold dimer L1L NF-YC3 | | Descriptor: | ACETATE ION, CALCIUM ION, NUCLEAR TRANSCRIPTION FACTOR Y SUBUNIT B-6, ... | | Authors: | Gnesutta, N, Saad, D, Chaves-Sanjuan, A, Mantovani, R, Nardini, M. | | Deposit date: | 2016-05-06 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Arabidopsis thaliana L1L/NF-YC3 Histone-fold Dimer Reveals Specificities of the LEC1 Family of NF-Y Subunits in Plants.
Mol Plant, 10, 2017
|
|
5GW9
 
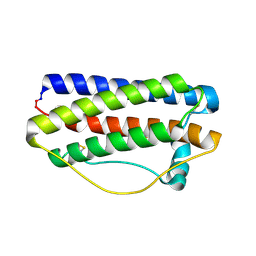 | | Crystal structure of C163, a backbone circularized G-CSF | | Descriptor: | Granulocyte colony-stimulating factor | | Authors: | Miyafusa, T, Honda, S. | | Deposit date: | 2016-09-09 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural insights into the backbone-circularized granulocyte colony-stimulating factor containing a short connector.
Biochem. Biophys. Res. Commun., 500, 2018
|
|
2WMB
 
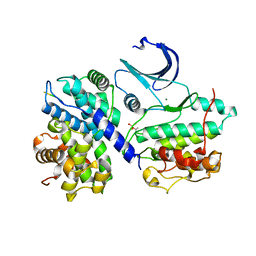 | | Structural and thermodynamic consequences of cyclization of peptide ligands for the recruitment site of cyclin A | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, CYCLIN-A2, LINEAR RKLFD, ... | | Authors: | Robertson, G.F, Endicott, J.A, Noble, M.E.M, McDonnell, J.M. | | Deposit date: | 2009-06-30 | | Release date: | 2010-09-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Thermodynamic Consequences of Cyclization of Peptide Ligands for the Recruitment Site of Cyclin A
To be Published
|
|
5GPD
 
 | |
5GRS
 
 | | Complex structure of the fission yeast SREBP-SCAP binding domains | | Descriptor: | Sterol regulatory element-binding protein 1, Sterol regulatory element-binding protein cleavage-activating protein | | Authors: | Gong, X, Qian, H.W, Wu, J.P, Yan, N. | | Deposit date: | 2016-08-12 | | Release date: | 2016-12-14 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Complex structure of the fission yeast SREBP-SCAP binding domains reveals an oligomeric organization
Cell Res., 26, 2016
|
|
6I3Y
 
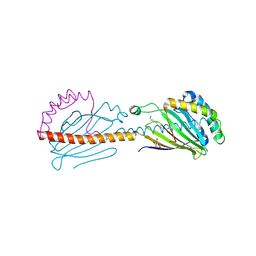 | | Crystal structure of the human mitochondrial PRELID1K58V-TRIAP1 complex with PS | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, O-[(R)-{[(2R)-2,3-bis(octadecanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-L-serine, PRELI domain-containing protein 1, ... | | Authors: | Miliara, X, Berry, J.-L, Morgan, R.M.L, Matthews, S.J. | | Deposit date: | 2018-11-08 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structural determinants of lipid specificity within Ups/PRELI lipid transfer proteins.
Nat Commun, 10, 2019
|
|
